Acute Lymphoblastic Leukaemia Cells Impair Dendritic Cell and Macrophage Differentiation: Role of BMP4
- PMID: 31337120
- PMCID: PMC6679123
- DOI: 10.3390/cells8070722
Acute Lymphoblastic Leukaemia Cells Impair Dendritic Cell and Macrophage Differentiation: Role of BMP4
Abstract
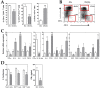
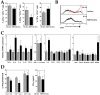
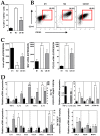
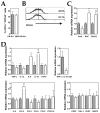
Dendritic cells and macrophages are common components of the tumour immune microenvironment and can contribute to immune suppression in both solid and haematological cancers. The Bone Morphogenetic Protein (BMP) pathway has been reported to be involved in cancer, and more recently in leukaemia development and progression. In the present study, we analyse whether acute lymphoblastic leukaemia (ALL) cells can affect the differentiation of dendritic cells and macrophages and the involvement of BMP pathway in the process. We show that ALL cells produce BMP4 and that conditioned media from ALL cells promote the generation of dendritic cells with immunosuppressive features and skew M1-like macrophage polarization towards a less pro-inflammatory phenotype. Likewise, BMP4 overexpression in ALL cells potentiates their ability to induce immunosuppressive dendritic cells and favours the generation of M2-like macrophages with pro-tumoral features. These results suggest that BMP4 is in part responsible for the alterations in dendritic cell and macrophage differentiation produced by ALL cells.
Keywords: BMP4; acute lymphoblastic leukaemia; dendritic cells; macrophages; tumour immune microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

