Value of Collaboration among Multi-Domain Experts in Analysis of High-Throughput Genomics Data
- PMID: 31337654
- PMCID: PMC6801074
- DOI: 10.1158/0008-5472.CAN-19-0769
Value of Collaboration among Multi-Domain Experts in Analysis of High-Throughput Genomics Data
Abstract
The recent explosion and ease of access to large-scale genomics data is intriguing. However, serious obstacles exist to the optimal management of the entire spectrum from data production in the laboratory through bioinformatic analysis to statistical evaluation and ultimately clinical interpretation. Beyond the multitude of technical issues, what stands out the most is the absence of adequate communication among the specialists in these domains. Successful interdisciplinary collaborations along the genomics pipeline extending from laboratory experiments to bioinformatic analyses to clinical application are notable in large scale, well managed projects such as The Cancer Genome Atlas. However, in certain settings in which the various experts perform their specialized research activities in isolation, the siloed approach to their research contributes to the generation of questionable genomic interpretations. Such situations are particularly concerning when the ultimate endpoint involves genetic/genomic interpretations that are intended for clinical applications. In spite of the fact that clinicians express interest in gaining a better understanding of clinical genomic applications, the lack of communication from upstream experts leaves them with a serious level of discomfort in applying such genomic knowledge to patient care. This discomfort is especially evident among healthcare providers who are not trained as geneticists, in particular primary care physicians. We offer some initiatives that have potential to address this problem, with emphasis on improved and ongoing communication among all the experts in these fields, constituting a comprehensive genomic "pipeline" from laboratory to patient.
©2019 American Association for Cancer Research.
Conflict of interest statement
“The authors declare no potential conflicts of interest.”
Figures
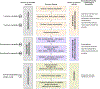
 Technical variability:
Technical variability:
 Use of fresh tissue versus FFPE (formalin fixed paraffin embedded) tissue; amount of input DNA
Use of fresh tissue versus FFPE (formalin fixed paraffin embedded) tissue; amount of input DNA Extent of DNA fragmentation
Extent of DNA fragmentation
 Bioinformatics variability-algorithm selection: (see
Bioinformatics variability-algorithm selection: (see  for abbreviation definitions)
for abbreviation definitions)
 Algorithm selection for alignment to reference sequence
Algorithm selection for alignment to reference sequence Algorithm selection for genetic alteration identification
Algorithm selection for genetic alteration identification
 Clinical interpretation of DNA motifs: SNVs (single nucleotide variants), SNPs (single nucleotide polymorphisms), VUS (variants of uncertain/unknown significance), INDELs (insertion/deletions), CNVs (copy number variants), fusion variants
Clinical interpretation of DNA motifs: SNVs (single nucleotide variants), SNPs (single nucleotide polymorphisms), VUS (variants of uncertain/unknown significance), INDELs (insertion/deletions), CNVs (copy number variants), fusion variants

 for abbreviation definitions
for abbreviation definitionsSimilar articles
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
-
The future of Cochrane Neonatal.Early Hum Dev. 2020 Nov;150:105191. doi: 10.1016/j.earlhumdev.2020.105191. Epub 2020 Sep 12. Early Hum Dev. 2020. PMID: 33036834
-
Integrating next-generation sequencing into pediatric oncology practice: An assessment of physician confidence and understanding of clinical genomics.Cancer. 2017 Jun 15;123(12):2352-2359. doi: 10.1002/cncr.30581. Epub 2017 Feb 13. Cancer. 2017. PMID: 28192596 Free PMC article.
-
Clinical application of genomic high-throughput data: Infrastructural, ethical, legal and psychosocial aspects.Eur Neuropsychopharmacol. 2020 Feb;31:1-15. doi: 10.1016/j.euroneuro.2019.09.008. Epub 2019 Dec 20. Eur Neuropsychopharmacol. 2020. PMID: 31866110 Review.
-
The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology: This report is based on a colloquium, “The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology,” sponsored by the American Academy of Microbiology and held October 11-13, 2002, in Longboat Key, Florida.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33119236 Free Books & Documents. Review.
Cited by
-
Editorial: Using Cancer 'Omics' to Understand Cancer.Front Oncol. 2020 Jul 24;10:1201. doi: 10.3389/fonc.2020.01201. eCollection 2020. Front Oncol. 2020. PMID: 32850349 Free PMC article. No abstract available.
-
Inference of Subpathway Activity Profiles Reveals Metabolism Abnormal Subpathway Regions in Glioblastoma Multiforme.Front Oncol. 2020 Sep 11;10:1549. doi: 10.3389/fonc.2020.01549. eCollection 2020. Front Oncol. 2020. PMID: 33072547 Free PMC article.
-
Humanizing Big Data: Recognizing the Human Aspect of Big Data.Front Oncol. 2020 Mar 13;10:186. doi: 10.3389/fonc.2020.00186. eCollection 2020. Front Oncol. 2020. PMID: 32231993 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

