Modeling the binding of diverse ligands within the Ah receptor ligand binding domain
- PMID: 31337850
- PMCID: PMC6650409
- DOI: 10.1038/s41598-019-47138-z
Modeling the binding of diverse ligands within the Ah receptor ligand binding domain
Abstract
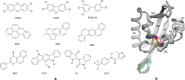
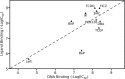
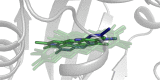
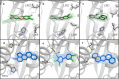
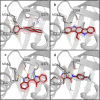
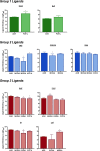
The Ah receptor (AhR) is a ligand-dependent transcription factor belonging to the basic helix-loop-helix Per-Arnt-Sim (bHLH-PAS) superfamily. Binding to and activation of the AhR by a variety of chemicals results in the induction of expression of diverse genes and production of a broad spectrum of biological and toxic effects. The AhR also plays important roles in several physiological responses, which has led it to become a novel target for the development of therapeutic drugs. Differences in the interactions of various ligands within the AhR ligand binding domain (LBD) may contribute to differential modulation of AhR functionality. We combined computational and experimental analyses to investigate the binding modes of a group of chemicals representative of major classes of AhR ligands. On the basis of a novel computational approach for molecular docking to the homology model of the AhR LBD that includes the receptor flexibility, we predicted specific residues within the AhR binding cavity that play a critical role in binding of three distinct groups of chemicals. The prediction was validated by site-directed mutagenesis and evaluation of the relative ligand binding affinities for the mutant AhRs. These results provide an avenue for understanding ligand modulation of the AhR functionality and for rational drug design.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
New aryl hydrocarbon receptor homology model targeted to improve docking reliability.J Chem Inf Model. 2011 Nov 28;51(11):2868-81. doi: 10.1021/ci2001617. Epub 2011 Nov 2. J Chem Inf Model. 2011. PMID: 21981577 Free PMC article.
-
Comparative In Vitro and In Silico Analysis of the Selectivity of Indirubin as a Human Ah Receptor Agonist.Int J Mol Sci. 2018 Sep 10;19(9):2692. doi: 10.3390/ijms19092692. Int J Mol Sci. 2018. PMID: 30201897 Free PMC article.
-
Detection of the TCDD binding-fingerprint within the Ah receptor ligand binding domain by structurally driven mutagenesis and functional analysis.Biochemistry. 2009 Jun 30;48(25):5972-83. doi: 10.1021/bi900259z. Biochemistry. 2009. PMID: 19456125 Free PMC article.
-
Ligand binding and activation of the Ah receptor.Chem Biol Interact. 2002 Sep 20;141(1-2):3-24. doi: 10.1016/s0009-2797(02)00063-7. Chem Biol Interact. 2002. PMID: 12213382 Review.
-
Mechanisms of ligand-induced aryl hydrocarbon receptor-mediated biochemical and toxic responses.Toxicol Pathol. 1998 Sep-Oct;26(5):657-71. doi: 10.1177/019262339802600510. Toxicol Pathol. 1998. PMID: 9789953 Review.
Cited by
-
Converging Roles of the Aryl Hydrocarbon Receptor in Early Embryonic Development, Maintenance of Stemness, and Tissue Repair.Toxicol Sci. 2021 Jul 16;182(1):1-9. doi: 10.1093/toxsci/kfab050. Toxicol Sci. 2021. PMID: 34009372 Free PMC article. Review.
-
Computational discovery of novel aryl hydrocarbon receptor modulators for psoriasis therapy.Sci Rep. 2025 Jun 6;15(1):19963. doi: 10.1038/s41598-025-03626-z. Sci Rep. 2025. PMID: 40481006 Free PMC article.
-
Metadynamics-Based Approaches for Modeling the Hypoxia-Inducible Factor 2α Ligand Binding Process.J Chem Theory Comput. 2021 Jul 13;17(7):3841-3851. doi: 10.1021/acs.jctc.1c00114. Epub 2021 Jun 3. J Chem Theory Comput. 2021. PMID: 34082524 Free PMC article.
-
Development, scrutiny, and modulation of transient reporter gene assays of the xenobiotic metabolism pathway in zebrafish hepatocytes.Cell Biol Toxicol. 2023 Jun;39(3):991-1013. doi: 10.1007/s10565-021-09659-0. Epub 2021 Oct 15. Cell Biol Toxicol. 2023. PMID: 34654992 Free PMC article.
-
6-Prenylnaringenin from Hops Disrupts ERα-Mediated Downregulation of CYP1A1 to Facilitate Estrogen Detoxification.Chem Res Toxicol. 2020 Nov 16;33(11):2793-2803. doi: 10.1021/acs.chemrestox.0c00194. Epub 2020 Oct 12. Chem Res Toxicol. 2020. PMID: 32986415 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

