Secondary Placental Defects in Cxadr Mutant Mice
- PMID: 31338035
- PMCID: PMC6628872
- DOI: 10.3389/fphys.2019.00622
Secondary Placental Defects in Cxadr Mutant Mice
Abstract
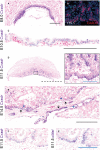
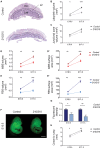
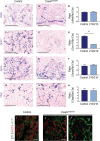
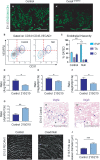
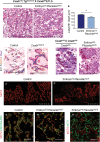
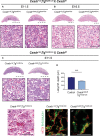
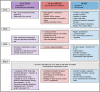
The Coxsackie virus and adenovirus receptor (CXADR) is an adhesion molecule known for its role in virus-cell interactions, epithelial integrity, and organogenesis. Loss of Cxadr causes numerous embryonic defects in mice, notably abnormal development of the cardiovascular system, and embryonic lethality. While CXADR expression has been reported in the placenta, the precise cellular localization and function within this tissue are unknown. Since impairments in placental development and function can cause secondary cardiovascular abnormalities, a phenomenon referred to as the placenta-heart axis, it is possible placental phenotypes in Cxadr mutant embryos may underlie the reported cardiovascular defects and embryonic lethality. In the current study, we determine the cellular localization of placental Cxadr expression and whether there are placental abnormalities in the absence of Cxadr. In the placenta, CXADR is expressed specifically by trophoblast labyrinth progenitors as well as cells of the visceral yolk sac (YS). In the absence of Cxadr, we observed altered expression of angiogenic factors coupled with poor expansion of trophoblast and fetal endothelial cell subpopulations, plus diminished placental transport. Unexpectedly, preserving endogenous trophoblast Cxadr expression revealed the placental defects to be secondary to primary embryonic and/or YS phenotypes. Moreover, further tissue-restricted deletions of Cxadr suggest that the secondary placental defects are likely influenced by embryonic lineages such as the fetal endothelium or those within the extraembryonic YS vascular plexus.
Keywords: Cxadr; fetal circulation; fetal heart; placenta; placenta-heart axis.
Figures
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

