The Type III Secretion System (T3SS)-Translocon of Atypical Enteropathogenic Escherichia coli (aEPEC) Can Mediate Adherence
- PMID: 31338081
- PMCID: PMC6629874
- DOI: 10.3389/fmicb.2019.01527
The Type III Secretion System (T3SS)-Translocon of Atypical Enteropathogenic Escherichia coli (aEPEC) Can Mediate Adherence
Abstract
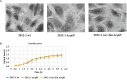
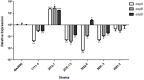
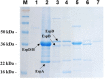
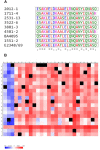
The intimin protein is the major adhesin involved in the intimate adherence of atypical enteropathogenic Escherichia coli (aEPEC) strains to epithelial cells, but little is known about the structures involved in their early colonization process. A previous study demonstrated that the type III secretion system (T3SS) plays an additional role in the adherence of an Escherichia albertii strain. Therefore, we assumed that the T3SS could be related to the adherence efficiency of aEPEC during the first stages of contact with epithelial cells. To test this hypothesis, we examined the adherence of seven aEPEC strains and their eae (intimin) isogenic mutants in the standard HeLa adherence assay and observed that all wild-type strains were adherent while five isogenic eae mutants were not. The two eae mutant strains that remained adherent were then used to generate the eae/escN double mutants (encoding intimin and the T3SS ATPase, respectively) and after the adherence assay, we observed that one strain lost its adherence capacity. This suggested a role for the T3SS in the initial adherence steps of this strain. In addition, we demonstrated that this strain expressed the T3SS at significantly higher levels when compared to the other wild-type strains and that it produced longer translocon-filaments. Our findings reveal that the T3SS-translocon can play an additional role as an adhesin at the beginning of the colonization process of aEPEC.
Keywords: EspA; EspB; EspD; adherence; atypical EPEC; gene expression; polymorphism; type III secretion system (T3SS)-translocon.
Figures


Similar articles
-
Dissection of the role of pili and type 2 and 3 secretion systems in adherence and biofilm formation of an atypical enteropathogenic Escherichia coli strain.Infect Immun. 2013 Oct;81(10):3793-802. doi: 10.1128/IAI.00620-13. Epub 2013 Jul 29. Infect Immun. 2013. PMID: 23897608 Free PMC article.
-
Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin.Microbiology (Reading). 2004 Mar;150(Pt 3):527-538. doi: 10.1099/mic.0.26740-0. Microbiology (Reading). 2004. PMID: 14993302
-
The Serine Protease EspC from Enteropathogenic Escherichia coli Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System.PLoS Pathog. 2015 Jul 1;11(7):e1005013. doi: 10.1371/journal.ppat.1005013. eCollection 2015 Jul. PLoS Pathog. 2015. PMID: 26132339 Free PMC article.
-
The localized adherence pattern of an atypical enteropathogenic Escherichia coli is mediated by intimin omicron and unexpectedly promotes HeLa cell invasion.Cell Microbiol. 2008 Feb;10(2):415-25. doi: 10.1111/j.1462-5822.2007.01054.x. Epub 2007 Oct 2. Cell Microbiol. 2008. PMID: 17910741
-
An overview of atypical enteropathogenic Escherichia coli.FEMS Microbiol Lett. 2009 Aug;297(2):137-49. doi: 10.1111/j.1574-6968.2009.01664.x. Epub 2009 May 27. FEMS Microbiol Lett. 2009. PMID: 19527295 Review.
Cited by
-
An outbreak associated with Escherichia albertii in a junior high school, China.Epidemiol Infect. 2024 Oct 4;152:e117. doi: 10.1017/S0950268824001341. Epidemiol Infect. 2024. PMID: 39363601 Free PMC article.
-
Virulence Potential of a Multidrug-Resistant Escherichia coli Strain Belonging to the Emerging Clonal Group ST101-B1 Isolated from Bloodstream Infection.Microorganisms. 2020 May 30;8(6):827. doi: 10.3390/microorganisms8060827. Microorganisms. 2020. PMID: 32486334 Free PMC article.
-
Plant-exuded chemical signals induce surface attachment of the bacterial pathogen Pseudomonas syringae.PeerJ. 2023 Mar 27;11:e14862. doi: 10.7717/peerj.14862. eCollection 2023. PeerJ. 2023. PMID: 37009160 Free PMC article.
-
Enzymatic Specificity of Conserved Rho GTPase Deamidases Promotes Invasion of Vibrio parahaemolyticus at the Expense of Infection.mBio. 2022 Aug 30;13(4):e0162922. doi: 10.1128/mbio.01629-22. Epub 2022 Jul 7. mBio. 2022. PMID: 35862776 Free PMC article.
-
Molecular Epidemiology and Presence of Hybrid Pathogenic Escherichia coli among Isolates from Community-Acquired Urinary Tract Infection.Microorganisms. 2022 Jan 27;10(2):302. doi: 10.3390/microorganisms10020302. Microorganisms. 2022. PMID: 35208757 Free PMC article.
References
-
- Abe C. M., Trabulsi L. R., Blanco J., Blanco M., Dahbi G., Blanco J. E., et al. (2009). Virulence features of atypical enteropathogenic Escherichia coli identified by the eae+ EAF-negative stx– genetic profile. Diagn. Microbiol. Infect. Dis. 64 357–365. 10.1016/j.diagmicrobio.2009.03.025 - DOI - PubMed
LinkOut - more resources
Full Text Sources

