Epigenetic Regulation at the Interplay Between Gut Microbiota and Host Metabolism
- PMID: 31338107
- PMCID: PMC6628876
- DOI: 10.3389/fgene.2019.00638
Epigenetic Regulation at the Interplay Between Gut Microbiota and Host Metabolism
Abstract
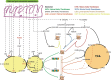
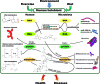
Gut microbiota communities have coevolved for millions of years in a symbiotic relationship with their mammalian hosts. Elucidating and understanding the molecular mechanisms by which microbiota interacts with its host and how this contributes to the homeostasis of the host is crucial. One of these molecular relationships is the so-called chemical crosstalk between microbiota and host metabolisms, including the poorly explored epigenetic regulation of host tissues by the metabolic activity of gut microbiota in response to changes in diet. DNA methylation and histone modifications are epigenetic marks partly regulated by enzymes such as methylases and acetylases, whose activity depend on host and microbiota metabolites that act as substrates and cofactors for these reactions. However, providing a complete mechanistic description of the regulatory interactions between both metabolisms and the impact on the expression of host genes through an epigenetic modulation, remains elusive. This article presents our perspective on how metabolomic, metagenomic, transcriptomic, and epigenomic data can be used to investigate the "microbiota-nutrient metabolism-epigenetics axis." We also discuss the implications and opportunities this knowledge may have for basic and applied science, such as the impact on the way we structure future research, understand, and prevent diseases like type 2 diabetes or obesity.
Keywords: epigenetics; histones; metabolism; metabolomics; microbiota; omics.
Figures
References
LinkOut - more resources
Full Text Sources

