Comammox Functionality Identified in Diverse Engineered Biological Wastewater Treatment Systems
- PMID: 31338378
- PMCID: PMC6650159
- DOI: 10.1021/acs.estlett.7b00577
Comammox Functionality Identified in Diverse Engineered Biological Wastewater Treatment Systems
Abstract
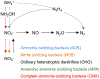
Complete ammonia oxidation (comammox) to nitrate by certain Nitrospira-lineage bacteria (CMX) could contribute to overall nitrogen cycling in engineered biological nitrogen removal (BNR) processes in addition to the more well-documented nitrogen transformations by ammonia-oxidizing bacteria (AOB), nitrite-oxidizing bacteria (NOB), and anaerobic ammonia-oxidizing (anammox) bacteria (AMX). A metagenomic survey was conducted to quantify the presence and elucidate the potential functionality of CMX in 16 full-scale BNR configurations treating mainstream or sidestream wastewater. CMX proposed to date were combined with previously published AOB, NOB, and AMX genomes to create an expanded database for alignment of metagenomic reads. CMX-assigned metagenomic reads accounted for between 0.28 and 0.64% of total coding DNA sequences in all BNR configurations. Phylogenetic analysis of key nitrification functional genes amoA, encoding the α-subunit of ammonia monooxygenase, haoB, encoding the β-subunit of hydroxylamine oxidoreductase, and nxrB, encoding the β-subunit of nitrite oxidoreductase, confirmed that each BNR system contained coding regions for production of these enzymes by CMX specifically. Ultimately, the ubiquitous presence of CMX bacteria and metabolic functionality in such diverse system configurations emphasizes the need to translate novel bacterial transformations to engineered biological process interrogation, operation, and design.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Grady CPL; Daigger GT; Love NG; Filipe CDM Biological Wastewater Treatment; IWA Publishing, 2011.
-
- Lu H; Chandran K; Stensel D Microbial ecology of denitrification in biological wastewater treatment. Water Res. 2014, 64, 237–254. - PubMed
-
- Hu B.-l.; Zheng P; Tang C.-j.; Chen J.-w.; van der Biezen E; Zhang L; Ni B.-j.; Jetten MS; Yan J; Yu H-Q; Kartal B. Identification and quantification of anammox bacteria in eight nitrogen removal reactors. Water Res. 2010, 44, 5014–5020. - PubMed
-
- Lackner S; Gilbert EM; Vlaeminck SE; Joss A; Horn H; van Loosdrecht MC Full-scale partial nitritation/anammox experiences–an application survey. Water Res. 2014, 55, 292–303. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
