Factors contributing to variability of glycan microarray binding profiles
- PMID: 31338503
- PMCID: PMC9335900
- DOI: 10.1039/c9fd00021f
Factors contributing to variability of glycan microarray binding profiles
Abstract
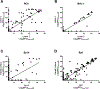
Protein-carbohydrate interactions play significant roles in a wide variety of biological systems. Glycan microarrays are commonly utilized to interrogate the selectivity, sensitivity, and breadth of these complex protein-carbohydrate interactions. During the past two decades, numerous distinct glycan microarray platforms have been developed, each assembled from a variety of slide-surface chemistries, glycan-attachment chemistries, glycan presentations, linkers, and glycan densities. Comparative analyses of glycan microarray data have shown that while many protein-carbohydrate interactions behave predictably across microarrays, there are instances when various array formats produce different results. For optimal construction and use of this technology, it is important to understand sources of variances across array platforms. In this study, we performed a systematic comparison of microarray data from 8 lectins across a range of concentrations on the CFG and neoglycoprotein array platforms. While there was good general agreement on the binding specificity of the lectins on the two arrays, there were some cases of large discrepancies. Differences in glycan density and linker composition contributed significantly to variability. The results provide insights for interpreting microarray data and designing future glycan microarrays.
Conflict of interest statement
Conflicts of interest
There are no conflicts to declare.
Figures

Similar articles
-
Use of glycan microarrays to explore specificity of glycan-binding proteins.Methods Enzymol. 2010;480:417-44. doi: 10.1016/S0076-6879(10)80033-3. Methods Enzymol. 2010. PMID: 20816220 Review.
-
Neoglycoprotein and Glycoprotein Printing on a Hydrogel Functionalized Microarray Surface and Incubation with Labeled Lectins.Methods Mol Biol. 2022;2460:3-24. doi: 10.1007/978-1-0716-2148-6_1. Methods Mol Biol. 2022. PMID: 34972927
-
General Strategies for Glycan Microarray Data Processing and Analysis.Methods Mol Biol. 2022;2460:67-87. doi: 10.1007/978-1-0716-2148-6_5. Methods Mol Biol. 2022. PMID: 34972931
-
Cross-platform comparison of glycan microarray formats.Glycobiology. 2014 Jun;24(6):507-17. doi: 10.1093/glycob/cwu019. Epub 2014 Mar 22. Glycobiology. 2014. PMID: 24658466 Free PMC article.
-
Glycan microarrays from construction to applications.Chem Soc Rev. 2022 Oct 3;51(19):8276-8299. doi: 10.1039/d2cs00452f. Chem Soc Rev. 2022. PMID: 36111958 Review.
Cited by
-
Carbohydrate antigen microarray analysis of serum IgG and IgM antibodies before and after adult porcine islet xenotransplantation in cynomolgus macaques.PLoS One. 2021 Jun 17;16(6):e0253029. doi: 10.1371/journal.pone.0253029. eCollection 2021. PLoS One. 2021. PMID: 34138941 Free PMC article.
-
Natural and Synthetic Sialylated Glycan Microarrays and Their Applications.Front Mol Biosci. 2019 Sep 13;6:88. doi: 10.3389/fmolb.2019.00088. eCollection 2019. Front Mol Biosci. 2019. PMID: 31572731 Free PMC article. Review.
-
The future of plant lectinology: Advanced technologies and computational tools.BBA Adv. 2025 Jan 28;7:100145. doi: 10.1016/j.bbadva.2025.100145. eCollection 2025. BBA Adv. 2025. PMID: 39958819 Free PMC article.
-
On-Chip Neo-Glycopeptide Synthesis for Multivalent Glycan Presentation.Chemistry. 2020 Aug 6;26(44):9954-9963. doi: 10.1002/chem.202001291. Epub 2020 Jun 11. Chemistry. 2020. PMID: 32315099 Free PMC article.
-
A Useful Guide to Lectin Binding: Machine-Learning Directed Annotation of 57 Unique Lectin Specificities.ACS Chem Biol. 2022 Nov 18;17(11):2993-3012. doi: 10.1021/acschembio.1c00689. Epub 2022 Jan 27. ACS Chem Biol. 2022. PMID: 35084820 Free PMC article.
References
-
- Yu AL; Gilman AL; Ozkaynak MF; London WB; Kreissman SG; Chen HX; Smith M; Anderson B; Villablanca JG; Matthay KK; Shimada H; Grupp SA; Seeger R; Reynolds CP; Buxton A; Reisfeld RA; Gillies SD; Cohn SL; Maris JM; Sondel PM Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N. Engl. J. Med 2010, 363, 1324–1334. - PMC - PubMed
-
- Cheung NKV; Cheung IY; Kushner BH; Ostrovnaya I; Chamberlain E; Kramer K; Modak S Murine anti-GD2 monoclonal antibody 3F8 combined with granulocyte- macrophage colony-stimulating factor and 13-cis-retinoic acid in high-risk patients with stage 4 neuroblastoma in first remission. J. Clin. Oncol 2012, 30, 3264–3270. - PMC - PubMed
-
- Geissner A; Seeberger PH Glycan Arrays: From Basic Biochemical Research to Bioanalytical and Biomedical Applications. Ann. Rev. Anal. Chem 2016, 9, 223–247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

