Microbial Experimental Evolution - a proving ground for evolutionary theory and a tool for discovery
- PMID: 31338963
- PMCID: PMC6680118
- DOI: 10.15252/embr.201846992
Microbial Experimental Evolution - a proving ground for evolutionary theory and a tool for discovery
Abstract
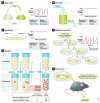
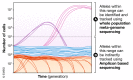
Microbial experimental evolution uses controlled laboratory populations to study the mechanisms of evolution. The molecular analysis of evolved populations enables empirical tests that can confirm the predictions of evolutionary theory, but can also lead to surprising discoveries. As with other fields in the life sciences, microbial experimental evolution has become a tool, deployed as part of the suite of techniques available to the molecular biologist. Here, I provide a review of the general findings of microbial experimental evolution, especially those relevant to molecular microbiologists that are new to the field. I also relate these results to design considerations for an evolution experiment and suggest future directions for those working at the intersection of experimental evolution and molecular biology.
Keywords: adaptation; directed evolution; experimental evolution; microbiome evolution; selection.
© 2019 The Author. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The author declares that he has no conflict of interest.
Figures

Similar articles
-
Genomic investigations of evolutionary dynamics and epistasis in microbial evolution experiments.Curr Opin Genet Dev. 2015 Dec;35:33-9. doi: 10.1016/j.gde.2015.08.008. Epub 2015 Sep 14. Curr Opin Genet Dev. 2015. PMID: 26370471 Free PMC article. Review.
-
Recombination Alters the Dynamics of Adaptation on Standing Variation in Laboratory Yeast Populations.Mol Biol Evol. 2018 Jan 1;35(1):180-201. doi: 10.1093/molbev/msx278. Mol Biol Evol. 2018. PMID: 29069452 Free PMC article.
-
Mechanistic approaches to the study of evolution: the functional synthesis.Nat Rev Genet. 2007 Sep;8(9):675-88. doi: 10.1038/nrg2160. Nat Rev Genet. 2007. PMID: 17703238 Free PMC article. Review.
-
Forecasting of phenotypic and genetic outcomes of experimental evolution in Pseudomonas protegens.PLoS Genet. 2021 Aug 5;17(8):e1009722. doi: 10.1371/journal.pgen.1009722. eCollection 2021 Aug. PLoS Genet. 2021. PMID: 34351900 Free PMC article.
-
The probability of evolutionary rescue: towards a quantitative comparison between theory and evolution experiments.Philos Trans R Soc Lond B Biol Sci. 2013 Jan 19;368(1610):20120088. doi: 10.1098/rstb.2012.0088. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23209169 Free PMC article.
Cited by
-
Frequent, infinitesimal bottlenecks maximize the rate of microbial adaptation.Genetics. 2023 Dec 6;225(4):iyad185. doi: 10.1093/genetics/iyad185. Genetics. 2023. PMID: 37804525 Free PMC article.
-
Assessing Rapid Adaptation Through Epigenetic Inheritance: A New Experimental Approach.Plant Cell Environ. 2025 Feb;48(2):1494-1499. doi: 10.1111/pce.15220. Epub 2024 Oct 25. Plant Cell Environ. 2025. PMID: 39450906 Free PMC article.
-
Unlocking the potential of experimental evolution to study drug resistance in pathogenic fungi.NPJ Antimicrob Resist. 2024 Dec 12;2(1):48. doi: 10.1038/s44259-024-00064-1. NPJ Antimicrob Resist. 2024. PMID: 39843963 Free PMC article. Review.
-
Evolving a plant-beneficial bacterium in soil vs. nutrient-rich liquid culture has contrasting effects on in-soil fitness.Appl Environ Microbiol. 2025 Apr 23;91(4):e0208524. doi: 10.1128/aem.02085-24. Epub 2025 Mar 11. Appl Environ Microbiol. 2025. PMID: 40067020 Free PMC article.
-
Adaptive laboratory evolution of microbial co-cultures for improved metabolite secretion.Mol Syst Biol. 2021 Aug;17(8):e10189. doi: 10.15252/msb.202010189. Mol Syst Biol. 2021. PMID: 34370382 Free PMC article.
References
-
- Garland T, Rose MR (2009) Experimental evolution: concepts, methods, and applications of selection experiments. Berkeley and Los Angeles, CA: University of California Press;
-
- Kassen R (2014) Experimental evolution and the nature of biodiversity. Greenwood Village, CO: Roberts and Company;
-
- Lenski RE, Rose MR, Simpson SC, Tadler SC (1991) Long‐term experimental evolution in Escherichia‐coli. I. Adaptation and divergence during 2,000 generations. Am Nat 138: 1315–1341
-
- Wiser MJ, Ribeck N, Lenski RE (2013) Long‐term dynamics of adaptation in asexual populations. Science 342: 1364–1367 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

