Rapid metagenomics analysis of EMS vehicles for monitoring pathogen load using nanopore DNA sequencing
- PMID: 31339905
- PMCID: PMC6655686
- DOI: 10.1371/journal.pone.0219961
Rapid metagenomics analysis of EMS vehicles for monitoring pathogen load using nanopore DNA sequencing
Abstract
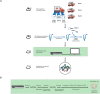
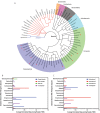
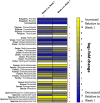
Pathogen monitoring, detection and removal are essential to public health and outbreak management. Systems are in place for monitoring the microbial load of hospitals and public health facilities with strategies to mitigate pathogen spread. However, no such strategies are in place for ambulances, which are tasked with transporting at-risk individuals in immunocompromised states. As standard culturing techniques require a laboratory setting, and are time consuming and labour intensive, our approach was designed to be portable, inexpensive and easy to use based on the MinION third-generation sequencing platform from Oxford Nanopore Technologies. We developed a transferable sampling-to-analysis pipeline to characterize the microbial community in emergency medical service vehicles. Our approach identified over sixty-eight organisms in ambulances to the genera level, with a proportion of these being connected with health-care associated infections, such as Clostridium spp. and Staphylococcus spp. We also monitored the microbiome of different locations across three ambulances over time, and examined the dynamic community of microorganisms found in emergency medical service vehicles. Observed differences identified hot spots, which may require heightened monitoring and extensive cleaning. Through metagenomics analysis it is also possible to identify how microorganisms spread between patients and colonize an ambulance over time. The sequencing results aid in the development of practices to mitigate disease spread, while also providing a useful tool for outbreak prediction through ongoing analysis of the ambulance microbiome to identify new and emerging pathogens. Overall, this pipeline allows for the tracking and monitoring of pathogenic microorganisms of epidemiological interest, including those related to health-care associated infections.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- World Health Organization. Managing meningitis epidemics in Africa: a quick reference guide for health authorities and health-care workers. Geneva, Switzerland: World Health Organization; 2015. - PubMed
-
- Weber DJ, Rutala WA, Miller MB, Huslage K, Sickbert-Bennett E. Role of hospital surfaces in the transmission of emerging health care-associated pathogens: norovirus, Clostridium difficile, and Acinetobacter species. Am J of Infect Control. 2010;38(5 Suppl 1):S25–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

