Molecular Mechanisms of p63-Mediated Squamous Cancer Pathogenesis
- PMID: 31340447
- PMCID: PMC6678256
- DOI: 10.3390/ijms20143590
Molecular Mechanisms of p63-Mediated Squamous Cancer Pathogenesis
Abstract
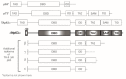
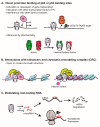
The p63 gene is a member of the p53/p63/p73 family of transcription factors and plays a critical role in development and homeostasis of squamous epithelium. p63 is transcribed as multiple isoforms; ΔNp63α, the predominant p63 isoform in stratified squamous epithelium, is localized to the basal cells and is overexpressed in squamous cell cancers of multiple organ sites, including skin, head and neck, and lung. Further, p63 is considered a stem cell marker, and within the epidermis, ΔNp63α directs lineage commitment. ΔNp63α has been implicated in numerous processes of skin biology that impact normal epidermal homeostasis and can contribute to squamous cancer pathogenesis by supporting proliferation and survival with roles in blocking terminal differentiation, apoptosis, and senescence, and influencing adhesion and migration. ΔNp63α overexpression may also influence the tissue microenvironment through remodeling of the extracellular matrix and vasculature, as well as by enhancing cytokine and chemokine secretion to recruit pro-inflammatory infiltrate. This review focuses on the role of ΔNp63α in normal epidermal biology and how dysregulation can contribute to cutaneous squamous cancer development, drawing from knowledge also gained by squamous cancers from other organ sites that share p63 overexpression as a defining feature.
Keywords: epidermal homeostasis; epidermal morphogenesis; keratinocytes; p53 family; p63; squamous carcinogenesis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Yang A., Kaghad M., Wang Y., Gillett E., Fleming M.D., Dotsch V., Andrews N.C., Caput D., McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/S1097-2765(00)80275-0. - DOI - PubMed
-
- Kaghad M., Bonnet H., Yang A., Creancier L., Biscan J.C., Valent A., Minty A., Chalon P., Lelias J.M., Dumont X., et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/S0092-8674(00)80540-1. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

