Ensemble of Deep Recurrent Neural Networks for Identifying Enhancers via Dinucleotide Physicochemical Properties
- PMID: 31340596
- PMCID: PMC6678823
- DOI: 10.3390/cells8070767
Ensemble of Deep Recurrent Neural Networks for Identifying Enhancers via Dinucleotide Physicochemical Properties
Abstract
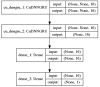
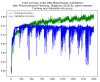
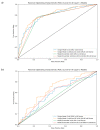
Enhancers are short deoxyribonucleic acid fragments that assume an important part in the genetic process of gene expression. Due to their possibly distant location relative to the gene that is acted upon, the identification of enhancers is difficult. There are many published works focused on identifying enhancers based on their sequence information, however, the resulting performance still requires improvements. Using deep learning methods, this study proposes a model ensemble of classifiers for predicting enhancers based on deep recurrent neural networks. The input features of deep ensemble networks were generated from six types of dinucleotide physicochemical properties, which had outperformed the other features. In summary, our model which used this ensemble approach could identify enhancers with achieved sensitivity of 75.5%, specificity of 76%, accuracy of 75.5%, and MCC of 0.51. For classifying enhancers into strong or weak sequences, our model reached sensitivity of 83.15%, specificity of 45.61%, accuracy of 68.49%, and MCC of 0.312. Compared to the benchmark result, our results had higher performance in term of most measurement metrics. The results showed that deep model ensembles hold the potential for improving on the best results achieved to date using shallow machine learning methods.
Keywords: biocomputing; dinucleotide physicochemical properties; enhancer DNA; ensemble deep learning; gene expression; high performance; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Rhie S.K., Tak Y.G., Guo Y., Yao L., Shen H., Coetzee G.A., Laird P.W., Farnham P.J. Identification of activated enhancers and linked transcription factors in breast, prostate, and kidney tumors by tracing enhancer networks using epigenetic traits. Epigenetics Chromatin. 2016;9:50. doi: 10.1186/s13072-016-0102-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

