The gut microbiota influences skeletal muscle mass and function in mice
- PMID: 31341063
- PMCID: PMC7501733
- DOI: 10.1126/scitranslmed.aan5662
The gut microbiota influences skeletal muscle mass and function in mice
Abstract
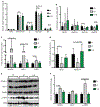
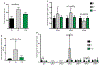
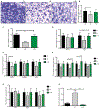
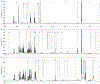
The functional interactions between the gut microbiota and the host are important for host physiology, homeostasis, and sustained health. We compared the skeletal muscle of germ-free mice that lacked a gut microbiota to the skeletal muscle of pathogen-free mice that had a gut microbiota. Compared to pathogen-free mouse skeletal muscle, germ-free mouse skeletal muscle showed atrophy, decreased expression of insulin-like growth factor 1, and reduced transcription of genes associated with skeletal muscle growth and mitochondrial function. Nuclear magnetic resonance spectrometry analysis of skeletal muscle, liver, and serum from germ-free mice revealed multiple changes in the amounts of amino acids, including glycine and alanine, compared to pathogen-free mice. Germ-free mice also showed reduced serum choline, the precursor of acetylcholine, the key neurotransmitter that signals between muscle and nerve at neuromuscular junctions. Reduced expression of genes encoding Rapsyn and Lrp4, two proteins important for neuromuscular junction assembly and function, was also observed in skeletal muscle from germ-free mice compared to pathogen-free mice. Transplanting the gut microbiota from pathogen-free mice into germ-free mice resulted in an increase in skeletal muscle mass, a reduction in muscle atrophy markers, improved oxidative metabolic capacity of the muscle, and elevated expression of the neuromuscular junction assembly genes Rapsyn and Lrp4 Treating germ-free mice with short-chain fatty acids (microbial metabolites) partly reversed skeletal muscle impairments. Our results suggest a role for the gut microbiota in regulating skeletal muscle mass and function in mice.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

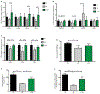
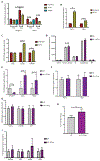
References
-
- Tintignac LA, Brenner H-R, Rüegg MA, Mechanisms regulating neuromuscular junction development and function and causes of muscle wasting. Physiol. Rev 95, 809–852 (2015). - PubMed
-
- Kelley DE, Goodpaster B, Wing RR, Simoneau JA, Skeletal muscle fatty acid metabolism in association with insulin resistance, obesity, and weight loss. Am. J. Physiol 277, E1130–E1141 (1999). - PubMed
-
- Nair KS, Aging muscle. Am. J. Clin. Nutr 81,953–963 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

