Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects
- PMID: 31341279
- PMCID: PMC6719697
- DOI: 10.1038/s41586-019-1414-x
Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects
Abstract
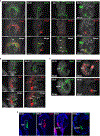
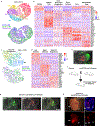
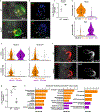
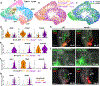
Organogenesis involves integration of diverse cell types; dysregulation of cell-type-specific gene networks results in birth defects, which affect 5% of live births. Congenital heart defects are the most common malformations, and result from disruption of discrete subsets of cardiac progenitor cells1, but the transcriptional changes in individual progenitors that lead to organ-level defects remain unknown. Here we used single-cell RNA sequencing to interrogate early cardiac progenitor cells as they become specified during normal and abnormal cardiogenesis, revealing how dysregulation of specific cellular subpopulations has catastrophic consequences. A network-based computational method for single-cell RNA-sequencing analysis that predicts lineage-specifying transcription factors2,3 identified Hand2 as a specifier of outflow tract cells but not right ventricular cells, despite the failure of right ventricular formation in Hand2-null mice4. Temporal single-cell-transcriptome analysis of Hand2-null embryos revealed failure of outflow tract myocardium specification, whereas right ventricular myocardium was specified but failed to properly differentiate and migrate. Loss of Hand2 also led to dysregulation of retinoic acid signalling and disruption of anterior-posterior patterning of cardiac progenitors. This work reveals transcriptional determinants that specify fate and differentiation in individual cardiac progenitor cells, and exposes mechanisms of disrupted cardiac development at single-cell resolution, providing a framework for investigating congenital heart defects.
Figures









References
-
- Srivastava D Making or breaking the heart: from lineage determination to morphogenesis. Cell 126, 1037–48 (2006). - PubMed
-
- Okawa S & del Sol A A computational strategy for predicting lineage specifiers in stem cell subpopulations. Stem Cell Res. 15, 427–434 (2015). - PubMed
-
- Srivastava D et al. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHand. Nat. Genet 16, 154–160 (1997). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

