De novo design of bioactive protein switches
- PMID: 31341284
- PMCID: PMC6733528
- DOI: 10.1038/s41586-019-1432-8
De novo design of bioactive protein switches
Abstract
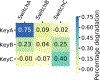
Allosteric regulation of protein function is widespread in biology, but is challenging for de novo protein design as it requires the explicit design of multiple states with comparable free energies. Here we explore the possibility of designing switchable protein systems de novo, through the modulation of competing inter- and intramolecular interactions. We design a static, five-helix 'cage' with a single interface that can interact either intramolecularly with a terminal 'latch' helix or intermolecularly with a peptide 'key'. Encoded on the latch are functional motifs for binding, degradation or nuclear export that function only when the key displaces the latch from the cage. We describe orthogonal cage-key systems that function in vitro, in yeast and in mammalian cells with up to 40-fold activation of function by key. The ability to design switchable protein functions that are controlled by induced conformational change is a milestone for de novo protein design, and opens up new avenues for synthetic biology and cell engineering.
Conflict of interest statement
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

