Transcriptome Analysis Reveals the Effect of Long Intergenic Noncoding RNAs on Pig Muscle Growth and Fat Deposition
- PMID: 31341893
- PMCID: PMC6614983
- DOI: 10.1155/2019/2951427
Transcriptome Analysis Reveals the Effect of Long Intergenic Noncoding RNAs on Pig Muscle Growth and Fat Deposition
Abstract
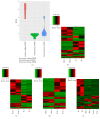
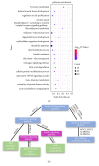
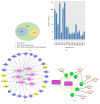
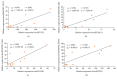
Muscle growth and fat deposition are the two important biological processes in the development of pigs which are closely related to the pig production performance. Long intergenic noncoding RNAs (lincRNAs), with lack of coding potential and the length of at least 200nt, have been extensively studied to play important roles in many biological processes. However, the importance and molecular regulation mechanism of lincRNAs in the process of muscle growth and fat deposition in pigs are still to be further studied comprehensively. In our study, we used the data, including liver, abdominal fat, and longissimus dorsi muscle of 240 days' age of two F2 full-sib female individuals from the white Duroc and Erhualian crossbreed, to identify 581 putative lincRNAs associated with pig muscle growth and fat deposition. The 581 putative lincRNAs shared many common features with other mammalian lincRNAs, such as fewer exons, lower expression levels, and shorter transcript lengths. Cross-tissue comparisons showed that many transcripts were tissue-specific and were involved in the important biological processes in their corresponding tissues. Gene ontology and pathway analysis revealed that many potential target genes (PTGs) of putative lincRNAs were involved in pig muscle growth and fat deposition-related processes, including muscle cell proliferation, lipid metabolism, and fatty acid degradation. In Quantitative Trait Locus (QTLs) analysis, some PTGs were screened from putative lincRNAs, MRPL12 is associated with muscle growth, GCGR and SLC25A10 were associated with fat deposition, and PPP3CA, DPYD, and FGGY were related not only to muscle growth but also to fat deposition. Therefore, it implied that these lincRNAs might participate in the biological processes related to muscle growth or fat deposition through homeostatic regulation of PTGs, but the detailed molecular regulatory mechanisms still needed to be further explored. This study lays the molecular foundation for the in-depth study of the role of lincRNAs in the pig muscle growth and fat deposition and further provides the new molecular markers for understanding the complex biological mechanisms of pig muscle growth and fat deposition.
Figures



References
-
- Duarte J. L., Cantet R. J., Rubio Y. L., et al. Refining genomewide association for growth and fat deposition traits in an F pig population. Journal of Animal Science. 2016;94(4):1387–1397. - PubMed
-
- Li P. H., Ma X., Zhang Y. Q., Zhang Q., Huang R. H. Progress in the physiological and genetic mechanisms underlying the high prolificacy of the Erhualian pig. Yi Chuan. 2017;39(11):1016–1024. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

