Past, present, and future of circRNAs
- PMID: 31343080
- PMCID: PMC6694216
- DOI: 10.15252/embj.2018100836
Past, present, and future of circRNAs
Abstract
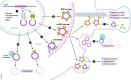
Exonic circular RNAs (circRNAs) are covalently closed RNA molecules generated by a process named back-splicing. circRNAs are highly abundant in eukaryotes, and many of them are evolutionary conserved. In metazoans, circular RNAs are expressed in a tissue-specific manner, are highly stable, and accumulate with age in neural tissues. circRNA biogenesis can regulate the production of the linear RNA counterpart in cis as back-splicing competes with linear splicing. Recent reports also demonstrate functions for some circRNAs in trans: Certain circRNAs interact with microRNAs, some are translated, and circRNAs have been shown to regulate immune responses and behavior. Here, we review current knowledge about animal circRNAs and summarize new insights into potential circRNA functions, concepts of their origin, and possible future directions in the field.
Keywords: RNA processing; circRNAs; circular RNAs; non-coding RNAs; splicing.
© 2019 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Aktas T, Avsar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R, Akhtar A (2017) DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544: 115–119 - PubMed
-
- Ashwal‐Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) CircRNA biogenesis competes with Pre‐mRNA splicing. Mol Cell 56: 55–66 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

