Alagille syndrome mutation update: Comprehensive overview of JAG1 and NOTCH2 mutation frequencies and insight into missense variant classification
- PMID: 31343788
- PMCID: PMC6899717
- DOI: 10.1002/humu.23879
Alagille syndrome mutation update: Comprehensive overview of JAG1 and NOTCH2 mutation frequencies and insight into missense variant classification
Abstract
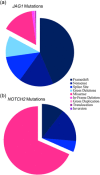
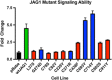
Alagille syndrome is an autosomal dominant disease with a known molecular etiology of dysfunctional Notch signaling caused primarily by pathogenic variants in JAGGED1 (JAG1), but also by variants in NOTCH2. The majority of JAG1 variants result in loss of function, however disease has also been attributed to lesser understood missense variants. Conversely, the majority of NOTCH2 variants are missense, though fewer of these variants have been described. In addition, there is a small group of patients with a clear clinical phenotype in the absence of a pathogenic variant. Here, we catalog our single-center study, which includes 401 probands and 111 affected family members amassed over a 27-year period, to provide updated mutation frequencies in JAG1 and NOTCH2 as well as functional validation of nine missense variants. Combining our cohort of 86 novel JAG1 and three novel NOTCH2 variants with previously published data (totaling 713 variants), we present the most comprehensive pathogenic variant overview for Alagille syndrome. Using this data set, we developed new guidance to help with the classification of JAG1 missense variants. Finally, we report clinically consistent cases for which a molecular etiology has not been identified and discuss the potential for next generation sequencing methodologies in novel variant discovery.
Keywords: Alagille syndrome; JAG1; NOTCH2; liver.
© 2019 The Authors. Human Mutation published by Wiley Periodicals, Inc.
Figures




References
-
- Alagille, D. , Estrada, A. , Hadchouel, M. , Gautler, M. , Odièvre, M. , & Dommergues, J. P. (1987). Syndromic paucity of interlobular bile ducts (Alagille syndrome or arteriohepatic dysplasia): Review of 80 cases. The Journal of Pediatrics, 110(2), 195–200. - PubMed
-
- Alagille, D. , Odièvre, M. , Gautier, M. , & Dommergues, J. P. (1975). Hepatic ductular hypoplasia associated with characteristic facies, vertebral malformations, retarded physical, mental, and sexual development, and cardiac murmur. The Journal of Pediatrics, 86(1), 63–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

