Reconstitution and Coupling of DNA Replication and Segregation in a Biomimetic System
- PMID: 31344304
- PMCID: PMC6899551
- DOI: 10.1002/cbic.201900299
Reconstitution and Coupling of DNA Replication and Segregation in a Biomimetic System
Abstract
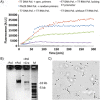
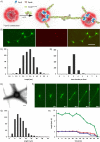
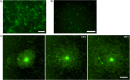
A biomimetic system capable of replication and segregation of genetic material constitutes an essential component for the future design of a minimal synthetic cell. Here we have used the simple T7 bacteriophage system and the plasmid-derived ParMRC system to establish in vitro DNA replication and DNA segregation, respectively. These processes were incorporated into biomimetic compartments providing an enclosed reaction space. The functional lifetime of the encapsulated segregation system could be prolonged by equipping it with ATP-regenerating and oxygen-scavenging systems. Finally, we showed that DNA replication and segregation processes could be coupled in vitro by using condensed DNA nanoparticles resulting from DNA replication. ParM spindles extended over tens of micrometers and could thus be used for segregation in compartments that are significantly longer than bacterial cell size. Overall, this work demonstrates the successful bottom-up assembly and coupling of molecular machines that mediate replication and segregation, thus providing an important step towards the development of a fully functional minimal cell.
Keywords: DNA nanoparticles; DNA replication; DNA segregation; ParM; T7; minimal cell.
© 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Figures



References
-
- Schwille P., Spatz J., Landfester K., Bodenschatz E., Herminghaus S., Sourjik V., Erb T. J., Bastiaens P., Lipowsky R., Hyman A., Dabrock P., Baret J.-C., Vidakovic-Koch T., Bieling P., Dimova R., Mutschler H., Robinson T., Tang T.-Y. D., Wegner S., Sundmacher K., Angew. Chem. Int. Ed. 2018, 57, 13382; - PubMed
- Angew. Chem. 2018, 130, 13566.
-
- Fischer H., Hinkle D. C., J. Biol. Chem. 1980, 255, 7956. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

