Phylogeography of Puumala orthohantavirus in Europe
- PMID: 31344894
- PMCID: PMC6723369
- DOI: 10.3390/v11080679
Phylogeography of Puumala orthohantavirus in Europe
Abstract
Puumala virus is an RNA virus hosted by the bank vole (Myodes glareolus) and is today present in most European countries. Whilst it is generally accepted that hantaviruses have been tightly co-evolving with their hosts, Puumala virus (PUUV) evolutionary history is still controversial and so far has not been studied at the whole European level. This study attempts to reconstruct the phylogeographical spread of modern PUUV throughout Europe during the last postglacial period in the light of an upgraded dataset of complete PUUV small (S) segment sequences and by using most recent computational approaches. Taking advantage of the knowledge on the past migrations of its host, we identified at least three potential independent dispersal routes of PUUV during postglacial recolonization of Europe by the bank vole. From the Alpe-Adrian region (Balkan, Austria, and Hungary) to Western European countries (Germany, France, Belgium, and Netherland), and South Scandinavia. From the vicinity of Carpathian Mountains to the Baltic countries and to Poland, Russia, and Finland. The dissemination towards Denmark and North Scandinavia is more hypothetical and probably involved several independent streams from south and north Fennoscandia.
Keywords: bank vole (myodes glareolus); co-evolution; phylogeography; puumala orthohantavirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
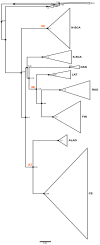


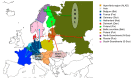
References
-
- Brummer-Korvenkontio M., Vaheri A., Hovi T., von Bonsdorff C.H., Vuorimies J., Manni T., Penttinen K., Oker-Blom N., Lahdevirta J. Nephropathia epidemica: Detection of antigen in bank voles and serologic diagnosis of human infection. J. Infect. Dis. 1980;141:131–134. doi: 10.1093/infdis/141.2.131. - DOI - PubMed
-
- Jaaskelainen K.M., Kaukinen P., Minskaya E.S., Plyusnina A., Vapalahti O., Elliott R.M., Weber F., Vaheri A., Plyusnin A. Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J. Med. Virol. 2007;79:1527–1536. doi: 10.1002/jmv.20948. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

