HuR interacts with lincBRN1a and lincBRN1b during neuronal stem cells differentiation
- PMID: 31345103
- PMCID: PMC6779397
- DOI: 10.1080/15476286.2019.1637698
HuR interacts with lincBRN1a and lincBRN1b during neuronal stem cells differentiation
Abstract
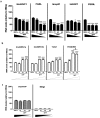
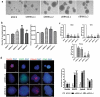
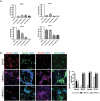
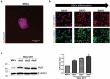
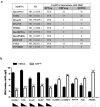
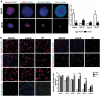
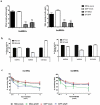
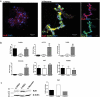
LncRNAs play crucial roles in cellular processes and their regulatory effects in the adult brain and neural stem cells (NSCs) remain to be entirely characterized. We report that 10 lncRNAs (LincENC1, FABL, lincp21, HAUNT, PERIL, lincBRN1a, lincBRN1b, HOTTIP, TUG1 and FENDRR) are expressed during murine NSCs differentiation and interact with the RNA-binding protein ELAVL1/HuR. Furthermore, we characterize the function of two of the deregulated lncRNAs, lincBRN1a and lincBRN1b, during NSCs' differentiation. Their inhibition leads to the induction of differentiation, with a concomitant decrease in stemness and an increase in neuronal markers, indicating that they exert key functions in neuronal cells differentiation. Furthermore, we describe here that HuR regulates their half-life, suggesting their synergic role in the differentiation process. We also identify six human homologs (PANTR1, TUG1, HOTTIP, TP53COR, ELDRR and FENDRR) of the mentioned 10 lncRNAs and we report their deregulation during human iPSCs differentiation into neurons. In conclusion, our results strongly indicate a key synergic role for lncRNAs and HuR in neuronal stem cells fate.
Keywords: HuR; Neural stem cell; differentiation; long-non-coding RNA.
Figures
References
-
- Temple S. Division and differentiation of isolated CNS blast cells in microculture. Nature. 1989;340(6233):471–473. - PubMed
-
- Rossi F, Cattaneo E. Opinion: neural stem cell therapy for neurological diseases: dreams and reality. Nat Rev Neurosci. 2002;3(5):401–409. - PubMed
-
- Carelli S, Giallongo T, Marfia G, et al. Exogenous adult postmortem neural precursors attenuate secondary degeneration and promote myelin sparing and functional recovery following experimental spinal cord injury. Cell Transplant. 2015;24(4):703–719. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
