Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study
- PMID: 31345709
- PMCID: PMC6715858
- DOI: 10.1016/S1473-3099(19)30392-5
Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study
Abstract
Background: A multidrug-resistant co-lineage of Plasmodium falciparum malaria, named KEL1/PLA1, spread across Cambodia in 2008-13, causing high rates of treatment failure with the frontline combination therapy dihydroartemisinin-piperaquine. Here, we report on the evolution and spread of KEL1/PLA1 in subsequent years.
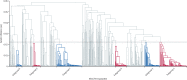
Methods: For this genomic epidemiology study, we analysed whole genome sequencing data from P falciparum clinical samples collected from patients with malaria between 2007 and 2018 from Cambodia, Laos, northeastern Thailand, and Vietnam, through the MalariaGEN P falciparum Community Project. Previously unpublished samples were provided by two large-scale multisite projects: the Tracking Artemisinin Resistance Collaboration II (TRAC2) and the Genetic Reconnaissance in the Greater Mekong Subregion (GenRe-Mekong) project. By investigating genome-wide relatedness between parasites, we inferred patterns of shared ancestry in the KEL1/PLA1 population.
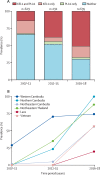
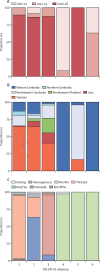
Findings: We analysed 1673 whole genome sequences that passed quality filters, and determined KEL1/PLA1 status in 1615. Before 2009, KEL1/PLA1 was only found in western Cambodia; by 2016-17 its prevalence had risen to higher than 50% in all of the surveyed countries except for Laos. In northeastern Thailand and Vietnam, KEL1/PLA1 exceeded 80% of the most recent P falciparum parasites. KEL1/PLA1 parasites maintained high genetic relatedness and low diversity, reflecting a recent common origin. Several subgroups of highly related parasites have recently emerged within this co-lineage, with diverse geographical distributions. The three largest of these subgroups (n=84, n=79, and n=47) mostly emerged since 2016 and were all present in Cambodia, Laos, and Vietnam. These expanding subgroups carried new mutations in the crt gene, which arose on a specific genetic background comprising multiple genomic regions. Four newly emerging crt mutations were rare in the early period and became more prevalent by 2016-17 (Thr93Ser, rising to 19·8%; His97Tyr to 11·2%; Phe145Ile to 5·5%; and Ile218Phe to 11·1%).
Interpretation: After emerging and circulating for several years within Cambodia, the P falciparum KEL1/PLA1 co-lineage diversified into multiple subgroups and acquired new genetic features, including novel crt mutations. These subgroups have rapidly spread into neighbouring countries, suggesting enhanced fitness. These findings highlight the urgent need for elimination of this increasingly drug-resistant parasite co-lineage, and the importance of genetic surveillance in accelerating malaria elimination efforts.
Funding: Wellcome Trust, Bill & Melinda Gates Foundation, UK Medical Research Council, and UK Department for International Development.
Copyright © 2019 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Figures

Comment in
-
Accelerated evolution and spread of multidrug-resistant Plasmodium falciparum takes down the latest first-line antimalarial drug in southeast Asia.Lancet Infect Dis. 2019 Sep;19(9):916-917. doi: 10.1016/S1473-3099(19)30394-9. Epub 2019 Jul 22. Lancet Infect Dis. 2019. PMID: 31345711 Free PMC article. No abstract available.
-
Plasmodium falciparum resistance to piperaquine driven by PfCRT.Lancet Infect Dis. 2019 Nov;19(11):1168-1169. doi: 10.1016/S1473-3099(19)30543-2. Lancet Infect Dis. 2019. PMID: 31657776 Free PMC article. No abstract available.
-
Multigenic architecture of piperaquine resistance trait in Plasmodium falciparum.Lancet Infect Dis. 2020 Jan;20(1):26-27. doi: 10.1016/S1473-3099(19)30689-9. Lancet Infect Dis. 2020. PMID: 31876497 No abstract available.
References
-
- Mita T, Tanabe K, Kita K. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol Int. 2009;58:201–209. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

