Distributing meiotic crossovers for optimal fertility and evolution
- PMID: 31345733
- PMCID: PMC6764855
- DOI: 10.1016/j.dnarep.2019.102648
Distributing meiotic crossovers for optimal fertility and evolution
Abstract
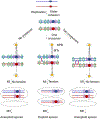
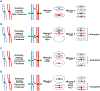
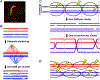
During meiosis, homologous chromosomes of a diploid cell are replicated and, without a second replication, are segregated during two nuclear divisions to produce four haploid cells (including discarded polar bodies in females of many species). Proper segregation of chromosomes at the first division requires in most species that homologous chromosomes be physically connected. Tension generated by connected chromosomes moving to opposite sides of the cell signals proper segregation. In the absence of the required connections, called crossovers, chromosomes often segregate randomly and produce aneuploid gametes and, thus, dead or disabled progeny. To be effective, crossovers must be properly distributed along chromosomes. Crossovers within or too near the centromere interfere with proper segregation; crossovers too near each other can ablate the required tension; and crossovers too concentrated in only one or a few regions would not re-assort most genetic characters important for evolution. Here, we discuss current knowledge of how the optimal distribution of crossovers is achieved in the fission yeast Schizosaccharomyces pombe, with reference to other well-studied species for comparison and illustration of the diversity of biology.
Keywords: Crossover interference; Crossover invariance; DNA break hotspots; DNA break interference; Homologous recombination; Linear element proteins; Meiosis; Pericentric repression; Schizosaccharomyces pombe.
Copyright © 2019 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures



References
-
- Powsner ER, Berman L, Number of chromosomes in the human cell, Nature, 188 (1960) 1045–1046. - PubMed
-
- Sinha B, Srivastava D, Jha J, Occurrence of various cytotypes of ophioglossum reticulatum L. in a population from N. E. India, Caryologia, 32 (1979) 135–146.
-
- Goday C, Pimpinelli S, Chromosome organization and heterochromatin elimination in parascaris, Science, 224 (1984) 411–413. - PubMed
-
- Crosland MW, Crazier RH, Myrmecia pilosula, an ant with only one pair of chromosomes, Science, 231 (1986) 1278. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

