Analysis of polygenic risk score usage and performance in diverse human populations
- PMID: 31346163
- PMCID: PMC6658471
- DOI: 10.1038/s41467-019-11112-0
Analysis of polygenic risk score usage and performance in diverse human populations
Abstract
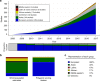
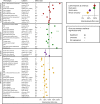
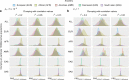
A historical tendency to use European ancestry samples hinders medical genetics research, including the use of polygenic scores, which are individual-level metrics of genetic risk. We analyze the first decade of polygenic scoring studies (2008-2017, inclusive), and find that 67% of studies included exclusively European ancestry participants and another 19% included only East Asian ancestry participants. Only 3.8% of studies were among cohorts of African, Hispanic, or Indigenous peoples. We find that predictive performance of European ancestry-derived polygenic scores is lower in non-European ancestry samples (e.g. African ancestry samples: t = -5.97, df = 24, p = 3.7 × 10-6), and we demonstrate the effects of methodological choices in polygenic score distributions for worldwide populations. These findings highlight the need for improved treatment of linkage disequilibrium and variant frequencies when applying polygenic scoring to cohorts of non-European ancestry, and bolster the rationale for large-scale GWAS in diverse human populations.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

