The Methods Employed in Mass Spectrometric Analysis of Posttranslational Modifications (PTMs) and Protein-Protein Interactions (PPIs)
- PMID: 31347048
- PMCID: PMC7059822
- DOI: 10.1007/978-3-030-15950-4_10
The Methods Employed in Mass Spectrometric Analysis of Posttranslational Modifications (PTMs) and Protein-Protein Interactions (PPIs)
Abstract
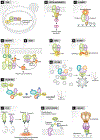
Mass Spectrometry (MS) has revolutionized the way we study biomolecules, especially proteins, their interactions and posttranslational modifications (PTM). As such MS has established itself as the leading tool for the analysis of PTMs mainly because this approach is highly sensitive, amenable to high throughput and is capable of assigning PTMs to specific sites in the amino acid sequence of proteins and peptides. Along with the advances in MS methodology there have been improvements in biochemical, genetic and cell biological approaches to mapping the interactome which are discussed with consideration for both the practical and technical considerations of these techniques. The interactome of a species is generally understood to represent the sum of all potential protein-protein interactions. There are still a number of barriers to the elucidation of the human interactome or any other species as physical contact between protein pairs that occur by selective molecular docking in a particular spatiotemporal biological context are not easily captured and measured.PTMs massively increase the complexity of organismal proteomes and play a role in almost all aspects of cell biology, allowing for fine-tuning of protein structure, function and localization. There are an estimated 300 PTMS with a predicted 5% of the eukaryotic genome coding for enzymes involved in protein modification, however we have not yet been able to reliably map PTM proteomes due to limitations in sample preparation, analytical techniques, data analysis, and the substoichiometric and transient nature of some PTMs. Improvements in proteomic and mass spectrometry methods, as well as sample preparation, have been exploited in a large number of proteome-wide surveys of PTMs in many different organisms. Here we focus on previously published global PTM proteome studies in the Apicomplexan parasites T. gondii and P. falciparum which offer numerous insights into the abundance and function of each of the studied PTM in the Apicomplexa. Integration of these datasets provide a more complete picture of the relative importance of PTM and crosstalk between them and how together PTM globally change the cellular biology of the Apicomplexan protozoa. A multitude of techniques used to investigate PTMs, mostly techniques in MS-based proteomics, are discussed for their ability to uncover relevant biological function.
Keywords: Cell cycle; Experimental techniques; In-silico databases; Mass spectrometry; Posttranslational crosstalk; Posttranslational modifications; Protein-protein interactions; Toxoplasma gondii.
Figures


References
-
- Brauch H; Kishida T; Glavac D; Chen F; Pausch F; Hofler H; Latif F; Lerman MI; Zbar B; Neumann HP, Von Hippel-Lindau (VHL) disease with pheochromocytoma in the Black Forest region of Germany: evidence for a founder effect. Human genetics 1995, 95 (5), 551–6. - PubMed
-
- Ohh M; Park CW; Ivan M; Hoffman MA; Kim TY; Huang LE; Pavletich N; Chau V; Kaelin WG, Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol 2000, 2 (7), 423–7. - PubMed
-
- Khare SD; Fleishman SJ, Emerging themes in the computational design of novel enzymes and protein-protein interfaces. FEBS Lett 2013, 587 (8), 1147–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

