DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation
- PMID: 31347257
- PMCID: PMC6726903
- DOI: 10.15252/embr.201847498
DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation
Abstract
A CGG trinucleotide repeat expansion in the 5' UTR of FMR1 causes the neurodegenerative disorder Fragile X-associated tremor/ataxia syndrome (FXTAS). This repeat supports a non-canonical mode of protein synthesis known as repeat-associated, non-AUG (RAN) translation. The mechanism underlying RAN translation at CGG repeats remains unclear. To identify modifiers of RAN translation and potential therapeutic targets, we performed a candidate-based screen of eukaryotic initiation factors and RNA helicases in cell-based assays and a Drosophila melanogaster model of FXTAS. We identified multiple modifiers of toxicity and RAN translation from an expanded CGG repeat in the context of the FMR1 5'UTR. These include the DEAD-box RNA helicase belle/DDX3X, the helicase accessory factors EIF4B/4H, and the start codon selectivity factors EIF1 and EIF5. Disrupting belle/DDX3X selectively inhibited FMR1 RAN translation in Drosophila in vivo and cultured human cells, and mitigated repeat-induced toxicity in Drosophila and primary rodent neurons. These findings implicate RNA secondary structure and start codon fidelity as critical elements mediating FMR1 RAN translation and identify potential targets for treating repeat-associated neurodegeneration.
Keywords: DDX3X; Fragile X-associated tremor/ataxia syndrome; RAN translation; RNA helicase; eIF.
© 2019 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
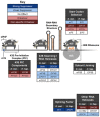

- A
Representative photographs of fly eyes expressing (CGG)90‐EGFP under a GMR‐GAL4 driver, with various belle disruptions.
- B
Quantitation of GMR‐GAL4 and (CGG)90‐EGFP eye phenotypes with belle disruptions (Mann–Whitney U‐test with Bonferroni corrections for multiple comparisons; n = 35–77/genotype).
- C, D
Longevity assays of (CGG)90‐EGFP; Tub5‐GS (log‐rank Mantel–Cox test with Bonferroni corrections for multiple comparisons; n = 110–219/genotype) and (CGG)90‐EGFP; ElaV‐GS (n = 147–299/genotype) flies with belle knockdown.
- E
Western blots of the FMRpolyG‐EGFP RAN product in (CGG)90‐EGFP; Tub5‐GS flies with and without belle knockdown by two independent shRNAs.
- F
Quantitation of FMRpolyG‐EGFP band density, normalized to β‐tubulin band density, from blots in (E) (Student's t‐test; n = 4–5/genotype).
- G
Abundance of (CGG)90‐EGFP mRNA normalized to RPL32 mRNA, following belle knockdown, determined by qRT–PCR (n = 8/genotype).
- H
Western blot of AUG‐driven EGFP in EGFP; Tub5‐GS flies with and without belle knockdown.
- I
Quantitation of EGFP band density, normalized to β‐tubulin band density, from blot in (H) (n = 4/genotype).
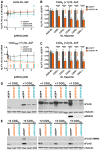
- A
Dose–response curves showing the effects of two independent anti‐DDX3X siRNAs on the expression of AUG‐NL‐3xF (top) and (CGG)100 +1 NL‐3xF (bottom) reporters. Plasmid‐based reporters were transfected into HeLa cells 24 h after knockdown, and reporter expression was quantified by luminescence. Nanoluciferase (NL) luminescence has been normalized to luminescence from firefly luciferase (FF), which was co‐transfected, in order to control for transfection variability. Asterisks refer to comparisons between anti‐DDX3X siRNAs and siRNAs against EGFP (siEGFP; two‐way ANOVA with Dunnett's multiple comparisons test; n = 12/condition).
- B, C
(CGG)n +1 and (CGG)n +2 NL‐3xF expression (normalized to FF) with and without DDX3X knockdown across a range of CGG repeat sizes. Black asterisks refer to comparisons between siDDX3X‐ and siEGFP‐treated cells; orange asterisks refer to comparisons between siDDX3X‐treated cells expressing AUG‐NL‐3xF and those expressing a different reporter (two‐way ANOVA with Tukey's multiple comparisons test; n = 17–30/condition).
- D, E
Western blots of FMRpolyG‐NL‐3xF and FMRpolyA‐NL‐3xF products with and without DDX3X knockdown across a range of repeat sizes.
The expression of in vitro‐transcribed AUG, +1 (CGG)100, and +2 (CGG)100 NL‐3xF RNAs following DDX3X knockdown in HeLa cells, expressed as NL luminescence normalized to FF luminescence. (Student's t‐test with Bonferroni corrections for multiple comparisons; n = 21/condition).
Abundance of reporter mRNAs following DDX3X knockdown and plasmid‐reporter transfection, determined by qRT–PCR (n = 7/condition). This panel depicts data as means ± SEM.
The expression of AUG‐NL‐3xF and +1 (CGG)100 NL‐3xF in in vitro translation extracts, collected from HeLa cells treated with siRNAs against EGFP or DDX3X (two‐way ANOVA with Tukey's multiple comparisons test; n = 4/condition).
Enrichment of HSPA1A and +1 (CGG)100 NL‐3xF mRNA following anti‐DDX3X RIP, relative to incubation with isotype control IgG. MALAT RNA, in contrast, is not enriched (Student's t‐test, n = 3). Data from the additional replicate are presented in Appendix Fig S8A.
The expression of +1 and +2 (CGG)100 NL‐3xF plasmid reporters with and without an AUG inserted 5′ to the CGG repeat, with and without DDX3X knockdown. Black asterisks refer to comparisons between siDDX3X‐ and siEGFP‐treated cells; orange asterisks refer to comparisons between siDDX3X‐treated cells expressing either +1 or +2 (CGG)100 NL‐3xF and those expressing the respective AUG‐driven variant (two‐way ANOVA with Tukey's multiple comparisons test; n = 11–12/condition).
The expression of NL‐3xF plasmids with initiator AUG codons mutated to near‐AUG codons, with and without DDX3X knockdown (two‐way ANOVA with Dunnett's multiple comparisons test; n = 18–24/condition). Black asterisks refer to comparisons between siEGFP‐treated and siDDX3X‐treated cells; orange asterisks refer to comparisons between siDDX3X‐treated cells expressing AUG‐NL‐3xF and those expressing a different reporter; white asterisks refer to comparisons between siDDX3X‐treated cells expressing +1 (CGG)100 NL‐3xF and those expressing a different reporter.
The expression of in vitro‐transcribed near‐AUG reporter RNAs in in vitro translation extracts, collected from HeLa cells treated with siRNAs against EGFP or DDX3X. Experiments with independent, replicate lysates are presented in Appendix Fig S7C (n = 4/group).

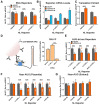
- A
Representative photographs of GMR‐GAL4; (CGG)90‐EGFP fly eyes expressing manipulations of eIF4B and eIF4H.
- B
Quantitation of GMR‐GAL4, (CGG)90‐EGFP eye phenotypes with eIF4B/H manipulations (Mann–Whitney U‐test with Bonferroni corrections for multiple comparisons; n = 26–55/genotype).
- C, D
The expression of plasmid‐based AUG‐NL and +1 (CGG)100 NL‐3xF reporters (C), or co‐transfected AUG‐FF reporters (D), following knockdown of EIF4B or EIF4H. Black asterisks refer to comparisons between siEGFP‐ and siEIF4B/H‐treated cells; pink and blue asterisks refer to comparisons between siEIF4B‐ (pink) or siEIF4H‐ (blue) treated cells expressing AUG‐NL‐3xF and those expressing +1 (CGG)100 (two‐way ANOVA with Tukey's multiple comparisons test, n = 9/condition).
- E
The expression of plasmid‐based AUG‐NL‐3xF and (CGG)100 +1 NL‐3xF reporters with and without over‐expression of EIF4B, EIF4H, or both (two‐way ANOVA with Dunnett's multiple comparisons test; n = 20/condition). Asterisks refer to comparisons between cells over‐expressing either EGFP or EIF4B, EIF4H, or EIF4B and EIF4H and expressing the same reporter.
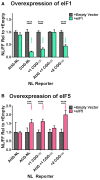
The expression of plasmid‐based NL‐3xF reporters in HEK293 cells with and without over‐expression of EIF1 (two‐way ANOVA with Sidak's multiple comparisons test; n = 9–12/condition). Black asterisks refer to comparisons between empty vector‐transfected and EIF1‐transfected cells; green asterisks refer to comparisons between EIF1‐transfected cells expressing AUG‐NL‐3xF and those expressing a different reporter.
The expression of plasmid‐based NL‐3xF reporters in HEK293 cells with and without over‐expression of EIF5 (two‐way ANOVA with Sidak's multiple comparisons test; n = 9–12/condition). Black asterisks refer to comparisons between empty vector‐transfected and EIF5‐transfected cells; pink asterisks refer to comparisons between EIF5‐transfected cells expressing AUG‐NL‐3xF and those expressing a different reporter.
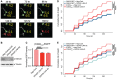
- A
Sample micrographs collected by automated longitudinal fluorescence microscopy, demonstrating the automated determination of cell death.
- B
Anti‐DDX3X Western blot of B35 cells transfected with either of two independent anti‐DDX3X LNAs or a control LNA.
- C
The expression of EGFP in primary rat neurons transfected with (CGG)100 (+1) EGFP and either anti‐DDX3X LNAs or a control (one‐way ANOVA with Tukey's multiple comparisons test; n = 2,408–5,689 cells/condition). All graphs depict pooled data, normalized first within the replicate.
- D, E
Transfection of anti‐DDX3X LNA #1 (D) or #2 (E) reduced the cumulative risk of death in (CGG)100 (+1) EGFP‐expressing neurons (Cox proportional hazard analysis; n = 2,408–3,676 cells/condition).
Similar articles
-
Repeat-associated non-AUG translation from antisense CCG repeats in fragile X tremor/ataxia syndrome.Ann Neurol. 2016 Dec;80(6):871-881. doi: 10.1002/ana.24800. Epub 2016 Nov 26. Ann Neurol. 2016. PMID: 27761921 Free PMC article.
-
CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome.Neuron. 2013 May 8;78(3):440-55. doi: 10.1016/j.neuron.2013.03.026. Epub 2013 Apr 18. Neuron. 2013. PMID: 23602499 Free PMC article.
-
Dissecting the roles of EIF4G homologs reveals DAP5 as a modifier of CGG repeat-associated toxicity in a Drosophila model of FXTAS.Neurobiol Dis. 2023 Aug;184:106212. doi: 10.1016/j.nbd.2023.106212. Epub 2023 Jun 22. Neurobiol Dis. 2023. PMID: 37352983 Free PMC article.
-
Between Order and Chaos: Understanding the Mechanism and Pathology of RAN Translation.Biol Pharm Bull. 2023;46(2):139-146. doi: 10.1248/bpb.b22-00448. Biol Pharm Bull. 2023. PMID: 36724941 Review.
-
Repeat-associated non-AUG (RAN) translation and other molecular mechanisms in Fragile X Tremor Ataxia Syndrome.Brain Res. 2018 Aug 15;1693(Pt A):43-54. doi: 10.1016/j.brainres.2018.02.006. Epub 2018 Feb 14. Brain Res. 2018. PMID: 29453961 Free PMC article. Review.
Cited by
-
Expansion of Clinical and Genetic Spectrum of DDX3X Neurodevelopmental Disorder in 23 Chinese Patients.Front Mol Neurosci. 2022 Mar 22;15:793001. doi: 10.3389/fnmol.2022.793001. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35392274 Free PMC article.
-
Mechanistic convergence across initiation sites for RAN translation in fragile X associated tremor ataxia syndrome.Hum Mol Genet. 2022 Jul 21;31(14):2317-2332. doi: 10.1093/hmg/ddab353. Hum Mol Genet. 2022. PMID: 35137065 Free PMC article.
-
Secondary structures that regulate mRNA translation provide insights for ASO-mediated modulation of cardiac hypertrophy.Nat Commun. 2023 Oct 3;14(1):6166. doi: 10.1038/s41467-023-41799-1. Nat Commun. 2023. PMID: 37789015 Free PMC article.
-
A novel de novo DDX3X missense variant in a female with brachycephaly and intellectual disability: a case report.Ital J Pediatr. 2021 Mar 31;47(1):81. doi: 10.1186/s13052-021-01033-4. Ital J Pediatr. 2021. PMID: 33789733 Free PMC article.
-
Non-canonical initiation factors modulate repeat-associated non-AUG translation.Hum Mol Genet. 2022 Aug 17;31(15):2521-2534. doi: 10.1093/hmg/ddac021. Hum Mol Genet. 2022. PMID: 35220421 Free PMC article.
References
-
- La Spada AR, Paulson HL, Fischbeck KH (1994) Trinucleotide repeat expansion in neurological disease. Ann Neurol 36: 814–822 - PubMed
-
- Paulson HL, Perez MK, Trottier Y, Trojanowski JQ, Subramony SH, Das SS, Vig P, Mandel JL, Fischbeck KH, Pittman RN (1997) Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 19: 333–344 - PubMed
-
- Warrick JM, Paulson HL, Gray‐Board GL, Bui QT, Fischbeck KH, Pittman RN, Bonini NM (1998) Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila . Cell 93: 939–949 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- T32GM007315/HHS | NIH | National Institute of General Medical Sciences (NIGMS)/International
- 1I01BX003231/U.S. Department of Veterans Affairs (VA)/International
- R01NS097542/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)/International
- I01 BX003231/BX/BLRD VA/United States
- T32 GM007863/GM/NIGMS NIH HHS/United States
- T32 NS007222/NS/NINDS NIH HHS/United States
- R01 NS099280/NS/NINDS NIH HHS/United States
- T32 GM007315/GM/NIGMS NIH HHS/United States
- P30 AG053760/AG/NIA NIH HHS/United States
- R01NS086810/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)/International
- 1P30AG053760/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)/International
- R01 NS097542/NS/NINDS NIH HHS/United States
- F31NS100302/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)/International
- R01 NS086810/NS/NINDS NIH HHS/United States
- 1I21BX001841/U.S. Department of Veterans Affairs (VA)/International
- T32NS007222/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)/International
- R01NS099280/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)/International
- F31 NS100302/NS/NINDS NIH HHS/United States
- F30 NS098571/NS/NINDS NIH HHS/United States
- I21 BX001841/BX/BLRD VA/United States
- F30NS098571/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)/International
- Michigan Alzheimer's Disease Center and Protein Folding Disease Initiative/International
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

