Synapse diversity and synaptome architecture in human genetic disorders
- PMID: 31348488
- PMCID: PMC6872429
- DOI: 10.1093/hmg/ddz178
Synapse diversity and synaptome architecture in human genetic disorders
Abstract
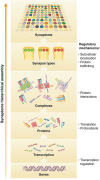
Over 130 brain diseases are caused by mutations that disrupt genes encoding the proteome of excitatory synapses. These include neurological and psychiatric disorders with early and late onset such as autism, schizophrenia and depression and many other rarer conditions. The proteome of synapses is highly complex with over 1000 conserved proteins which are differentially expressed generating a vast, potentially unlimited, number of synapse types. The diversity of synapses and their location in the brain are described by the synaptome. A recent study has mapped the synaptome across the mouse brain, revealing that synapse diversity is distributed into an anatomical architecture observed at scales from individual dendrites to the whole systems level. The synaptome architecture is built from the hierarchical expression and assembly of proteins into complexes and supercomplexes which are distributed into different synapses. Mutations in synapse proteins change the synaptome architecture leading to behavioral phenotypes. Mutations in the mechanisms regulating the hierarchical assembly of the synaptome, including transcription and proteostasis, may also change synapse diversity and synaptome architecture. The logic of synaptome hierarchical assembly provides a mechanistic framework that explains how diverse genetic disorders can converge on synapses in different brain circuits to produce behavioral phenotypes.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Husi H., Ward M.A., Choudhary J.S., Blackstock W.P. and Grant S.G. (2000) Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci., 3, 661–669. - PubMed
-
- Husi H. and Grant S.G. (2001) Isolation of 2000-kDa complexes of N-methyl-D-aspartate receptor and postsynaptic density 95 from mouse brain. J. Neurochem., 77, 281–291. - PubMed

