Diverse and dynamic DNA modifications in brain and diseases
- PMID: 31348493
- PMCID: PMC6872432
- DOI: 10.1093/hmg/ddz179
Diverse and dynamic DNA modifications in brain and diseases
Abstract
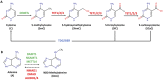
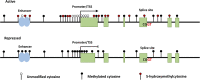
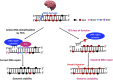
DNA methylation is a class of epigenetic modification essential for coordinating gene expression timing and magnitude throughout normal brain development and for proper brain function following development. Aberrant methylation changes are associated with changes in chromatin architecture, transcriptional alterations and a host of neurological disorders and diseases. This review highlights recent advances in our understanding of the methylome's functionality and covers potential new roles for DNA methylation, their readers, writers, and erasers. Additionally, we examine novel insights into the relationship between the methylome, DNA-protein interactions, and their contribution to neurodegenerative diseases. Lastly, we outline the gaps in our knowledge that will likely be filled through the widespread use of newer technologies that provide greater resolution into how individual cell types are affected by disease and the contribution of each individual modification site to disease pathogenicity.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
References
-
- Grassi D., Franz H., Vezzali R., Bovio P., Heidrich S., Dehghanian F., Lagunas N., Belzung C., Krieglstein K. and Vogel T. (2017) Neuronal activity, TGFβ-signaling and unpredictable chronic stress modulate transcription of Gadd45 family members and DNA methylation in the hippocampus. Cereb. Cortex, 27, 4166–4181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

