Metagenomic analysis of the effects of toll-like receptors on bacterial infection in the peritoneal cavity following cecum ligation and puncture in mice
- PMID: 31348811
- PMCID: PMC6660085
- DOI: 10.1371/journal.pone.0220398
Metagenomic analysis of the effects of toll-like receptors on bacterial infection in the peritoneal cavity following cecum ligation and puncture in mice
Abstract
Objective: To establish the composition of bacteria in mice following cecum ligation and puncture (CLP) through metagenomic analysis and investigate the role of TLRs on the composition of bacteria.
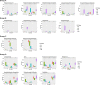
Methods: Total DNA extraction was done from the ascites, blood, and fecal samples from C57BL/6 mice sacrificed at 0, 4, 8, and 16 h, as well as from Tlr2-/-, Tlr4-/-, Tlr5-/-, and NF-κB-/-mice sacrificed at 16 h following CLP. Amplification of the V3-V4 regions of the bacterial 16S rRNA genes by PCR and the Illumina MiSeq sequencer was used for deep sequencing. Hierarchical clustering of the isolates was performed with Ward's method using Euclidean distances. The relative abundance according to operational taxonomic unit (OTU) number or taxa was used to compare the richness among subgroups in the experiments.
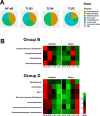
Results: There were 18 taxa that had significantly different abundances among the different samples of the C57BL/6 mice at 16 h following CLP. Various dynamic changes in the infectious bacteria inside the peritoneal cavity after CLP were found. While knockout of Tlr5 and NF-κB impaired the ability of bacterial clearance inside the peritoneal cavity for some kinds of bacteria found in the C57BL/6 mice, the knockout of Tlr4 enhanced clearance for other kinds of bacteria, and they presented excessive abundance in the peritoneal cavity despite their scarce abundance in the stool.
Conclusion: NF-κB and TLRs are involved in bacterial clearance and in the expression pattern of the bacterial abundance inside the peritoneal cavity during polymicrobial infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Magnotti LJ, Deitch EA. Burns, bacterial translocation, gut barrier function, and failure. J Burn Care Rehabil. 2005;26(5):383–91. Epub 2005/09/10. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

