Radiosensitivity Differences between EGFR Mutant and Wild-Type Lung Cancer Cells are Larger at Lower Doses
- PMID: 31349558
- PMCID: PMC6696360
- DOI: 10.3390/ijms20153635
Radiosensitivity Differences between EGFR Mutant and Wild-Type Lung Cancer Cells are Larger at Lower Doses
Abstract
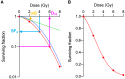
In the era of precision medicine, radiotherapy strategies should be determined based on genetic profiles that predict tumor radiosensitivity. Accordingly, pre-clinical research aimed at discovering clinically applicable genetic profiles is needed. However, how a given genetic profile affects cancer cell radiosensitivity is unclear. To address this issue, we performed a pilot in vitro study by utilizing EGFR mutational status as a model for genetic profile. Clonogenic assays of EGFR mutant (n = 6) and wild-type (n = 9) non-small cell lung carcinoma (NSCLC) cell lines were performed independently by two oncologists. Clonogenic survival parameters SF2, SF4, SF6, SF8, mean inactivation dose (MID), D10, D50, α, and β were obtained using the linear quadratic model. The differences in the clonogenic survival parameters between the EGFR mutant and wild-type cell lines were assessed using the Mann-Whitney U test. As a result, for both datasets, the p values for SF2, SF4, D50, α, and α/β were below 0.05, and those for SF2 were lowest. These data indicate that a genetic profile of NSCLC cell lines might be predictive for their radiation response; i.e., EGFR mutant cell lines might be more sensitive to low dose- and low fraction sized-irradiation.
Keywords: clonogenic assays; gene mutations; precision medicine; radiation therapy; radiosensitivity.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Jordan E.J., Kim H.R., Arcila M.E., Barron D., Chakravarty D., Gao J., Chang M.T., Ni A., Kundra R., Jonsson P., et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7:596–609. doi: 10.1158/2159-8290.CD-16-1337. - DOI - PMC - PubMed
-
- Lamba J.K., Chauhan L., Shin M., Loken M.R., Pollard J.A., Wang Y.C., Ries R.E., Aplenc R., Hirsch B.A., Raimondi S.C., et al. CD33 Splicing Polymorphism Determines Gemtuzumab Ozogamicin Response in De Novo Acute Myeloid Leukemia: Report from Randomized Phase III Children’s Oncology Group Trial AAML0531. J. Clin. Oncol. 2017;35:2674–2682. doi: 10.1200/JCO.2016.71.2513. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

