Unusual Organization of I-BAR Proteins on Tubular and Vesicular Membranes
- PMID: 31349990
- PMCID: PMC6697384
- DOI: 10.1016/j.bpj.2019.06.025
Unusual Organization of I-BAR Proteins on Tubular and Vesicular Membranes
Abstract
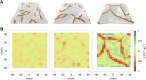
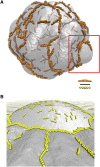
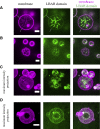
Protein-mediated membrane remodeling is a ubiquitous and critical process for proper cellular function. Inverse Bin/Amphiphysin/Rvs (I-BAR) domains drive local membrane deformation as a precursor to large-scale membrane remodeling. We employ a multiscale approach to provide the molecular mechanism of unusual I-BAR domain-driven membrane remodeling at a low protein surface concentration with near-atomistic detail. We generate a bottom-up coarse-grained model that demonstrates similar membrane-bound I-BAR domain aggregation behavior as our recent Mesoscopic Membrane with Explicit Proteins model. Together, these models bridge several length scales and reveal an aggregation behavior of I-BAR domains. We find that at low surface coverage (i.e., low bound protein density), I-BAR domains form transient, tip-to-tip strings on periodic flat membrane sheets. Inside of lipid bilayer tubules, we find linear aggregates parallel to the axis of the tubule. Finally, we find that I-BAR domains form tip-to-tip aggregates around the edges of membrane domes. These results are supported by in vitro experiments showing low curvature bulges surrounded by I-BAR domains on giant unilamellar vesicles. Overall, our models reveal new I-BAR domain aggregation behavior in membrane tubules and on the surface of vesicles at low surface concentration that add insight into how I-BAR domain proteins may contribute to certain aspects of membrane remodeling in cells.
Copyright © 2019 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

