Alveolar macrophages generate a noncanonical NRF2-driven transcriptional response to Mycobacterium tuberculosis in vivo
- PMID: 31350281
- PMCID: PMC6910245
- DOI: 10.1126/sciimmunol.aaw6693
Alveolar macrophages generate a noncanonical NRF2-driven transcriptional response to Mycobacterium tuberculosis in vivo
Abstract
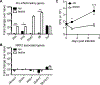
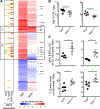
Alveolar macrophages (AMs) are the first cells to be infected during Mycobacterium tuberculosis (M.tb.) infection. Thus, the AM response to infection is the first of many steps leading to initiation of the adaptive immune response required for efficient control of infection. A hallmark of M.tb. infection is the slow initiation of the adaptive response, yet the mechanisms responsible for this are largely unknown. To study the initial AM response to infection, we developed a system to identify, sort, and analyze M.tb.-infected AMs from the lung within the first 10 days of infection. In contrast to what has been previously described using in vitro systems, M.tb.-infected AMs up-regulate a cell-protective antioxidant transcriptional signature that is dependent on the lung environment but not bacterial virulence. Computational approaches including pathway analysis and transcription factor motif enrichment analysis identify NRF2 as a master regulator of the response. Using knockout mouse models, we demonstrate that NRF2 drives expression of the cell-protective signature in AMs and impairs the control of early bacterial growth. AMs up-regulate a substantial pro-inflammatory response to M.tb. infection only 10 days after infection, yet comparisons with bystander AMs from the same infected animals demonstrate that M.tb.-infected AMs generate a less robust inflammatory response than the uninfected cells around them. Our findings demonstrate that the initial macrophage response to M.tb. in the lung is far less inflammatory than has previously been described by in vitro systems and may impede the overall host response to infection.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures





References
-
- Reid MJA, Arinaminpathy N, Bloom A, Bloom BR, Boehme C, Chaisson R, Chin DP, Churchyard G, Cox H, Ditiu L, Dybul M, Farrar J, Fauci AS, Fekadu E, Fujiwara PI, Hallett TB, Hanson CL, Harrington M, Herbert N, Hopewell PC, Ikeda C, Jamison DT, Khan AJ, Koek I, Krishnan N, Motsoaledi A, Pai M, Raviglione MC, Sharman A, Small PM, Swaminathan S, Temesgen Z, Vassall A, Venkatesan N, van Weezenbeek K, Yamey G, Agins BD, Alexandru S, Andrews JR, Beyeler N, Bivol S, Brigden G, Cattamanchi A, Cazabon D, Crudu V, Daftary A, Dewan P, Doepel LK, Eisinger RW, Fan V, Fewer S, Furin J, Goldhaber-Fiebert JD, Gomez GB, Graham SM, Gupta D, Kamene M, Khaparde S, Mailu EW, Masini EO, McHugh L, Mitchell E, Moon S, Osberg M, Pande T, Prince L, Rade K, Rao R, Remme M, Seddon JA, Selwyn C, Shete P, Sachdeva KS, Stallworthy G, Vesga JF, Vilc V, Goosby EP, Building a tuberculosis-free world: The Lancet Commission on tuberculosis. Lancet 393, 1331–1384 (2019). - PubMed
-
- Wolf AJ, Linas B, Trevejo-Nunez GJ, Kincaid E, Tamura T, Takatsu K, Ernst JD, Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol 179, 2509–2519 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

