Plantibacter flavus, Curtobacterium herbarum, Paenibacillus taichungensis, and Rhizobium selenitireducens Endophytes Provide Host-Specific Growth Promotion of Arabidopsis thaliana, Basil, Lettuce, and Bok Choy Plants
- PMID: 31350315
- PMCID: PMC6752021
- DOI: 10.1128/AEM.00383-19
Plantibacter flavus, Curtobacterium herbarum, Paenibacillus taichungensis, and Rhizobium selenitireducens Endophytes Provide Host-Specific Growth Promotion of Arabidopsis thaliana, Basil, Lettuce, and Bok Choy Plants
Abstract
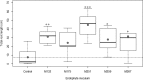
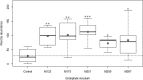
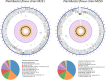
A collection of bacterial endophytes isolated from stem tissues of plants growing in soils highly contaminated with petroleum hydrocarbons were screened for plant growth-promoting capabilities. Twenty-seven endophytic isolates significantly improved the growth of Arabidopsis thaliana plants in comparison to that of uninoculated control plants. The five most beneficial isolates, one strain each of Curtobacterium herbarum, Paenibacillus taichungensis, and Rhizobium selenitireducens and two strains of Plantibacter flavus were further examined for growth promotion in Arabidopsis, lettuce, basil, and bok choy plants. Host-specific plant growth promotion was observed when plants were inoculated with the five bacterial strains. P. flavus strain M251 increased the total biomass and total root length of Arabidopsis plants by 4.7 and 5.8 times, respectively, over that of control plants and improved lettuce and basil root growth, while P. flavus strain M259 promoted Arabidopsis shoot and root growth, lettuce and basil root growth, and bok choy shoot growth. A genome comparison between P. flavus strains M251 and M259 showed that both genomes contain up to 70 actinobacterial putative plant-associated genes and genes involved in known plant-beneficial pathways, such as those for auxin and cytokinin biosynthesis and 1-aminocyclopropane-1-carboxylate deaminase production. This study provides evidence of direct plant growth promotion by Plantibacter flavusIMPORTANCE The discovery of new plant growth-promoting bacteria is necessary for the continued development of biofertilizers, which are environmentally friendly and cost-efficient alternatives to conventional chemical fertilizers. Biofertilizer effects on plant growth can be inconsistent due to the complexity of plant-microbe interactions, as the same bacteria can be beneficial to the growth of some plant species and neutral or detrimental to others. We examined a set of bacterial endophytes isolated from plants growing in a unique petroleum-contaminated environment to discover plant growth-promoting bacteria. We show that strains of Plantibacter flavus exhibit strain-specific plant growth-promoting effects on four different plant species.
Keywords: Arabidopsis; Plantibacter flavus; basil; bok choy; endophyte; host specificity; lettuce; plant growth promotion; plant microbiology.
Copyright © 2019 American Society for Microbiology.
Figures





Similar articles
-
Streptomyces Endophytes Promote Host Health and Enhance Growth across Plant Species.Appl Environ Microbiol. 2020 Aug 3;86(16):e01053-20. doi: 10.1128/AEM.01053-20. Print 2020 Aug 3. Appl Environ Microbiol. 2020. PMID: 32561579 Free PMC article.
-
Priming of Plant Growth Promotion by Volatiles of Root-Associated Microbacterium spp.Appl Environ Microbiol. 2018 Oct 30;84(22):e01865-18. doi: 10.1128/AEM.01865-18. Print 2018 Nov 15. Appl Environ Microbiol. 2018. PMID: 30194105 Free PMC article.
-
Isolation, identification and plant growth promotion ability of endophytic bacteria associated with lupine root nodule grown in Tunisian soil.Arch Microbiol. 2019 Dec;201(10):1333-1349. doi: 10.1007/s00203-019-01702-3. Epub 2019 Jul 15. Arch Microbiol. 2019. PMID: 31309236
-
Drought tolerance improvement in plants: an endophytic bacterial approach.Appl Microbiol Biotechnol. 2019 Sep;103(18):7385-7397. doi: 10.1007/s00253-019-10045-4. Epub 2019 Aug 2. Appl Microbiol Biotechnol. 2019. PMID: 31375881 Review.
-
Interaction of endophytic microbes with legumes.J Basic Microbiol. 2012 Jun;52(3):248-60. doi: 10.1002/jobm.201100063. Epub 2011 Sep 23. J Basic Microbiol. 2012. PMID: 21953403 Review.
Cited by
-
Domestication Impacts the Wheat-Associated Microbiota and the Rhizosphere Colonization by Seed- and Soil-Originated Microbiomes, Across Different Fields.Front Plant Sci. 2022 Jan 12;12:806915. doi: 10.3389/fpls.2021.806915. eCollection 2021. Front Plant Sci. 2022. PMID: 35095978 Free PMC article.
-
Unveiling the complete genome sequence of Paenibacillus taichungensis: genomic features and biocontrol potential.BMC Genom Data. 2025 Mar 17;26(1):18. doi: 10.1186/s12863-025-01310-9. BMC Genom Data. 2025. PMID: 40097926 Free PMC article.
-
Rhizospheric, seed, and root endophytic-associated bacteria of drought-tolerant legumes grown in arid soils of Namibia.Heliyon. 2024 Aug 24;10(17):e36718. doi: 10.1016/j.heliyon.2024.e36718. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39281591 Free PMC article.
-
Biocontrol and plant growth promoting traits of two avocado rhizobacteria are orchestrated by the emission of diffusible and volatile compounds.Front Microbiol. 2023 May 3;14:1152597. doi: 10.3389/fmicb.2023.1152597. eCollection 2023. Front Microbiol. 2023. PMID: 37206331 Free PMC article.
-
Diversity of maize (Zea mays L.) rhizobacteria with potential to promote plant growth.Braz J Microbiol. 2021 Dec;52(4):1807-1823. doi: 10.1007/s42770-021-00596-y. Epub 2021 Aug 30. Braz J Microbiol. 2021. PMID: 34458975 Free PMC article.
References
-
- Theis KR, Dheilly NM, Klassen JL, Brucker RM, Baines JF, Bosch TCG, Cryan JF, Gilbert SF, Goodnight CJ, Lloyd EA, Sapp J, Vandenkoornhuyse P, Zilber-Rosenberg I, Rosenberg E, Bordenstein SR. 2016. Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiomes. mSystems 1:e00028-16. doi: 10.1128/mSystems.00028-16. - DOI - PMC - PubMed
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

