Mosquito-Borne Viruses and Insect-Specific Viruses Revealed in Field-Collected Mosquitoes by a Monitoring Tool Adapted from a Microbial Detection Array
- PMID: 31350319
- PMCID: PMC6752009
- DOI: 10.1128/AEM.01202-19
Mosquito-Borne Viruses and Insect-Specific Viruses Revealed in Field-Collected Mosquitoes by a Monitoring Tool Adapted from a Microbial Detection Array
Abstract
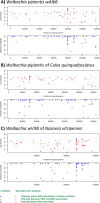
Several mosquito-borne diseases affecting humans are emerging or reemerging in the United States. The early detection of pathogens in mosquito populations is essential to prevent and control the spread of these diseases. In this study, we tested the potential applicability of the Lawrence Livermore Microbial Detection Array (LLMDA) to enhance biosurveillance by detecting microbes present in Aedes aegypti, Aedes albopictus, and Culex mosquitoes, which are major vector species globally, including in Texas. The sensitivity and reproducibility of the LLMDA were tested in mosquito samples spiked with different concentrations of dengue virus (DENV), revealing a detection limit of >100 but <1,000 PFU/ml. Additionally, field-collected mosquitoes from Chicago, IL, and College Station, TX, of known infection status (West Nile virus [WNV] and Culex flavivirus [CxFLAV] positive) were tested on the LLMDA to confirm its efficiency. Mosquito field samples of unknown infection status, collected in San Antonio, TX, and the Lower Rio Grande Valley (LRGV), TX, were run on the LLMDA and further confirmed by PCR or quantitative PCR (qPCR). The analysis of the field samples with the LLMDA revealed the presence of cell-fusing agent virus (CFAV) in A. aegypti populations. Wolbachia was also detected in several of the field samples (A. albopictus and Culex spp.) by the LLMDA. Our findings demonstrated that the LLMDA can be used to detect multiple arboviruses of public health importance, including viruses that belong to the Flavivirus, Alphavirus, and Orthobunyavirus genera. Additionally, insect-specific viruses and bacteria were also detected in field-collected mosquitoes. Another strength of this array is its ability to detect multiple viruses in the same mosquito pool, allowing for the detection of cocirculating pathogens in an area and the identification of potential ecological associations between different viruses. This array can aid in the biosurveillance of mosquito-borne viruses circulating in specific geographical areas.IMPORTANCE Viruses associated with mosquitoes have made a large impact on public and veterinary health. In the United States, several viruses, including WNV, DENV, and chikungunya virus (CHIKV), are responsible for human disease. From 2015 to 2018, imported Zika cases were reported in the United States, and in 2016 to 2017, local Zika transmission occurred in the states of Texas and Florida. With globalization and a changing climate, the frequency of outbreaks linked to arboviruses will increase, revealing a need to better detect viruses in vector populations. With the capacity of the LLMDA to detect viruses, bacteria, and fungi, this study highlights its ability to broadly screen field-collected mosquitoes and contribute to the surveillance and management of arboviral diseases.
Keywords: Aedes aegypti; Aedes albopictus; Culex; Culex flavivirus; Wolbachia; cell-fusing agent virus; insect-specific virus; microarrays.
Copyright © 2019 American Society for Microbiology.
Figures

References
-
- Kraemer MU, Sinka ME, Duda KA, Mylne A, Shearer FM, Brady OJ, Messina JP, Barker CM, Moore CG, Carvalho RG, Coelho GE, Van Bortel W, Hendrickx G, Schaffner F, Wint GR, Elyazar IR, Teng HJ, Hay SI. 2015. The global compendium of Aedes aegypti and Ae. albopictus occurrence. Sci Data 2:150035. doi: 10.1038/sdata.2015.35. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

