The AMPK-Related Kinases SIK1 and SIK3 Mediate Key Tumor-Suppressive Effects of LKB1 in NSCLC
- PMID: 31350328
- PMCID: PMC6825547
- DOI: 10.1158/2159-8290.CD-18-1261
The AMPK-Related Kinases SIK1 and SIK3 Mediate Key Tumor-Suppressive Effects of LKB1 in NSCLC
Abstract
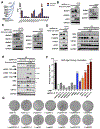
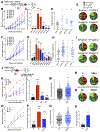
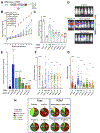
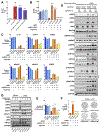
Mutations in the LKB1 (also known as STK11) tumor suppressor are the third most frequent genetic alteration in non-small cell lung cancer (NSCLC). LKB1 encodes a serine/threonine kinase that directly phosphorylates and activates 14 AMPK family kinases ("AMPKRs"). The function of many of the AMPKRs remains obscure, and which are most critical to the tumor-suppressive function of LKB1 remains unknown. Here, we combine CRISPR and genetic analysis of the AMPKR family in NSCLC cell lines and mouse models, revealing a surprising critical role for the SIK subfamily. Conditional genetic loss of Sik1 revealed increased tumor growth in mouse models of Kras-dependent lung cancer, which was further enhanced by loss of the related kinase Sik3. As most known substrates of the SIKs control transcription, gene-expression analysis was performed, revealing upregulation of AP1 and IL6 signaling in common between LKB1- and SIK1/3-deficient tumors. The SIK substrate CRTC2 was required for this effect, as well as for proliferation benefits from SIK loss. SIGNIFICANCE: The tumor suppressor LKB1/STK11 encodes a serine/threonine kinase frequently inactivated in NSCLC. LKB1 activates 14 downstream kinases in the AMPK family controlling growth and metabolism, although which kinases are critical for LKB1 tumor-suppressor function has remained an enigma. Here we unexpectedly found that two understudied kinases, SIK1 and SIK3, are critical targets in lung cancer.This article is highlighted in the In This Issue feature, p. 1469.
©2019 American Association for Cancer Research.
Conflict of interest statement
Competing Financial Interests
None
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

