1k-RiCA (1K-Rice Custom Amplicon) a novel genotyping amplicon-based SNP assay for genetics and breeding applications in rice
- PMID: 31350673
- PMCID: PMC6660535
- DOI: 10.1186/s12284-019-0311-0
1k-RiCA (1K-Rice Custom Amplicon) a novel genotyping amplicon-based SNP assay for genetics and breeding applications in rice
Abstract
Background: While a multitude of genotyping platforms have been developed for rice, the majority of them have not been optimized for breeding where cost, turnaround time, throughput and ease of use, relative to density and informativeness are critical parameters of their utility. With that in mind we report the development of the 1K-Rice Custom Amplicon, or 1k-RiCA, a robust custom sequencing-based amplicon panel of ~ 1000-SNPs that are uniformly distributed across the rice genome, designed to be highly informative within indica rice breeding pools, and tailored for genomic prediction in elite indica rice breeding programs.
Results: Empirical validation tests performed on the 1k-RiCA showed average marker call rates of 95% with marker repeatability and concordance rates of 99%. These technical properties were not affected when two common DNA extraction protocols were used. The average distance between SNPs in the 1k-RiCA was 1.5 cM, similar to the theoretical distance which would be expected between 1,000 uniformly distributed markers across the rice genome. The average minor allele frequencies on a panel of indica lines was 0.36 and polymorphic SNPs estimated on pairwise comparisons between indica by indica accessions and indica by japonica accessions were on average 430 and 450 respectively. The specific design parameters of the 1k-RiCA allow for a detailed view of genetic relationships and unambiguous molecular IDs within indica accessions and good cost vs. marker-density balance for genomic prediction applications in elite indica germplasm. Predictive abilities of Genomic Selection models for flowering time, grain yield, and plant height were on average 0.71, 0.36, and 0.65 respectively based on cross-validation analysis. Furthermore the inclusion of important trait markers associated with 11 different genes and QTL adds value to parental selection in crossing schemes and marker-assisted selection in forward breeding applications.
Conclusions: This study validated the marker quality and robustness of the 1k-RiCA genotypic platform for genotyping populations derived from indica rice subpopulation for genetic and breeding purposes including MAS and genomic selection. The 1k-RiCA has proven to be an alternative cost-effective genotyping system for breeding applications.
Keywords: Amplicon-based next generation sequencing; Breeding and genotyping; Genomic selection; Indica; Marker-assisted selection (MAS); Oryza sativa; SNP fingerprinting; Single nucleotide polymorphism (SNP).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

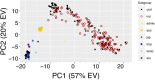


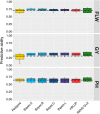
References
-
- Arbelaez JD, Moreno LT, Singh N, Tung CW, Maron LG, Ospina Y, Martinez CP, Grenier C, Lorieux M, McCouch S (2015) Development and GBS-genotyping of introgression lines (ILs) using two wild species of rice, O. meridionalis and O. rufipogon, in a common recurrent parent, O. sativa cv. Curinga. Mol breed 35:81. 10.1007/s11032-015-0276-7 - PMC - PubMed
-
- Bandillo N, Raghavan C, Muyco P, Sevilla MAL, Lobina IT, Dilla-Ermita C, Tung C-W, McCouch S, Thomson M, Mauleon R, Singh R, Gregorio G, Redoña E, Leung H. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice. 2013;6:11. doi: 10.1186/1939-8433-6-11. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

