Differentially methylated regions in bipolar disorder and suicide
- PMID: 31350827
- PMCID: PMC8375453
- DOI: 10.1002/ajmg.b.32754
Differentially methylated regions in bipolar disorder and suicide
Abstract
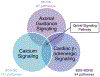
The addition of a methyl group to, typically, a cytosine-guanine dinucleotide (CpG) creates distinct DNA methylation patterns across the genome that can regulate gene expression. Aberrant DNA methylation of CpG sites has been associated with many psychiatric disorders including bipolar disorder (BD) and suicide. Using the SureSelectXT system, Methyl-Seq, we investigated the DNA methylation status of CpG sites throughout the genome in 50 BD individuals (23 subjects who died by suicide and 27 subjects who died from other causes) and 31 nonpsychiatric controls. We identified differentially methylated regions (DMRs) from three analyses: (a) BD subjects compared to nonpsychiatric controls (BD-NC), (b) BD subjects who died by suicide compared to nonpsychiatric controls (BDS-NC), and (c) BDS subjects compared to BD subjects who died from other causes (BDS-BDNS). One DMR from the BDS-NC analysis, located in ARHGEF38, was significantly hypomethylated (23.4%) in BDS subjects. This finding remained significant after multiple testing (PBootstrapped = 9.0 × 10-3 ), was validated using pyrosequencing, and was more significant in males. A secondary analysis utilized Ingenuity Pathway Analysis to identify enrichment in nominally significant DMRs. This identified an association with several pathways including axonal guidance signaling, calcium signaling, β-adrenergic signaling, and opioid signaling. Our comprehensive study provides further support that DNA methylation alterations influence the risk for BD and suicide. However, further investigation is required to confirm these associations and identify their functional consequences.
Keywords: ARHGEF38; opioid signaling; suicidal behavior; β-adrenergic signaling.
© 2019 Wiley Periodicals, Inc.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Figures

References
-
- Abdolmaleky HM, Gower AC, Wong CK, Cox JW, Zhang X, Thiagalingam A, … Thiagalingam S (2018). Aberrant transcriptomes and DNA methylomes define pathways that drive pathogenesis and loss of brain laterality/asymmetry in schizophrenia and bipolar disorder. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 180, 138–149. 10.1002/ajmg.b.32691 - DOI - PMC - PubMed

