Arrestin-3 interaction with maternal embryonic leucine-zipper kinase
- PMID: 31352007
- PMCID: PMC6717526
- DOI: 10.1016/j.cellsig.2019.109366
Arrestin-3 interaction with maternal embryonic leucine-zipper kinase
Abstract
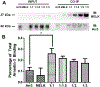
Maternal embryonic leucine-zipper kinase (MELK) overexpression impacts survival and proliferation of multiple cancer types, most notably glioblastomas and breast cancer. This makes MELK an attractive molecular target for cancer therapy. Yet the molecular mechanisms underlying the involvement of MELK in tumorigenic processes are unknown. MELK participates in numerous protein-protein interactions that affect cell cycle, proliferation, apoptosis, and embryonic development. Here we used both in vitro and in-cell assays to identify a direct interaction between MELK and arrestin-3. A part of this interaction involves the MELK kinase domain, and we further show that the interaction between the MELK kinase domain and arrestin-3 decreases the number of cells in S-phase, as compared to cells expressing the MELK kinase domain alone. Thus, we describe a new mechanism of regulation of MELK function, which may contribute to the control of cell fate.
Keywords: Arrestin; Cell fate signaling; Maternal embryonic leucine-zipper kinase (MELK); Protein-protein interactions.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures





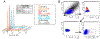
Similar articles
-
Maternal embryonic leucine zipper kinase (MELK): a novel regulator in cell cycle control, embryonic development, and cancer.Int J Mol Sci. 2013 Oct 31;14(11):21551-60. doi: 10.3390/ijms141121551. Int J Mol Sci. 2013. PMID: 24185907 Free PMC article. Review.
-
Involvement of maternal embryonic leucine zipper kinase (MELK) in mammary carcinogenesis through interaction with Bcl-G, a pro-apoptotic member of the Bcl-2 family.Breast Cancer Res. 2007;9(1):R17. doi: 10.1186/bcr1650. Breast Cancer Res. 2007. PMID: 17280616 Free PMC article.
-
Crystal structure of Maternal Embryonic Leucine Zipper Kinase (MELK) in complex with dorsomorphin (Compound C).Arch Biochem Biophys. 2019 Aug 15;671:1-7. doi: 10.1016/j.abb.2019.05.014. Epub 2019 May 17. Arch Biochem Biophys. 2019. PMID: 31108049
-
Maternal embryonic leucine zipper kinase in tumor cells and tumor microenvironment: An emerging player and promising therapeutic opportunity.Cancer Lett. 2023 Apr 28;560:216126. doi: 10.1016/j.canlet.2023.216126. Epub 2023 Mar 16. Cancer Lett. 2023. PMID: 36933780 Review.
-
Structural basis for the regulation of maternal embryonic leucine zipper kinase.PLoS One. 2013 Jul 26;8(7):e70031. doi: 10.1371/journal.pone.0070031. Print 2013. PLoS One. 2013. PMID: 23922895 Free PMC article.
Cited by
-
Arrestins: A Small Family of Multi-Functional Proteins.Int J Mol Sci. 2024 Jun 6;25(11):6284. doi: 10.3390/ijms25116284. Int J Mol Sci. 2024. PMID: 38892473 Free PMC article. Review.
-
β-Arrestins: Structure, Function, Physiology, and Pharmacological Perspectives.Pharmacol Rev. 2023 Sep;75(5):854-884. doi: 10.1124/pharmrev.121.000302. Epub 2023 Apr 7. Pharmacol Rev. 2023. PMID: 37028945 Free PMC article. Review.
-
GPCR-dependent and -independent arrestin signaling.Trends Pharmacol Sci. 2024 Jul;45(7):639-650. doi: 10.1016/j.tips.2024.05.007. Epub 2024 Jun 20. Trends Pharmacol Sci. 2024. PMID: 38906769 Free PMC article. Review.
-
β-arrestin2: an emerging player and potential therapeutic target in inflammatory immune diseases.Acta Pharmacol Sin. 2025 Sep;46(9):2347-2362. doi: 10.1038/s41401-024-01390-w. Epub 2024 Sep 30. Acta Pharmacol Sin. 2025. PMID: 39349766 Review.
-
Structure and function of β-arrestins, their emerging role in breast cancer, and potential opportunities for therapeutic manipulation.Adv Cancer Res. 2020;145:139-156. doi: 10.1016/bs.acr.2020.01.001. Epub 2020 Feb 5. Adv Cancer Res. 2020. PMID: 32089163 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

