Structure of the Human Core Centromeric Nucleosome Complex
- PMID: 31353180
- PMCID: PMC6702948
- DOI: 10.1016/j.cub.2019.06.062
Structure of the Human Core Centromeric Nucleosome Complex
Abstract
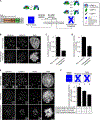
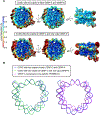
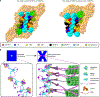
Centromeric nucleosomes are at the interface of the chromosome and the kinetochore that connects to spindle microtubules in mitosis. The core centromeric nucleosome complex (CCNC) harbors the histone H3 variant, CENP-A, and its binding proteins, CENP-C (through its central domain; CD) and CENP-N (through its N-terminal domain; NT). CENP-C can engage nucleosomes through two domains: the CD and the CENP-C motif (CM). CENP-CCD is part of the CCNC by virtue of its high specificity for CENP-A nucleosomes and ability to stabilize CENP-A at the centromere. CENP-CCM is thought to engage a neighboring nucleosome, either one containing conventional H3 or CENP-A, and a crystal structure of a nucleosome complex containing two copies of CENP-CCM was reported. Recent structures containing a single copy of CENP-NNT bound to the CENP-A nucleosome in the absence of CENP-C were reported. Here, we find that one copy of CENP-N is lost for every two copies of CENP-C on centromeric chromatin just prior to kinetochore formation. We present the structures of symmetric and asymmetric forms of the CCNC that vary in CENP-N stoichiometry. Our structures explain how the central domain of CENP-C achieves its high specificity for CENP-A nucleosomes and how CENP-C and CENP-N sandwich the histone H4 tail. The natural centromeric DNA path in our structures corresponds to symmetric surfaces for CCNC assembly, deviating from what is observed in prior structures using artificial sequences. At mitosis, we propose that CCNC asymmetry accommodates its asymmetric connections at the chromosome/kinetochore interface. VIDEO ABSTRACT.
Keywords: cell division; centromere; chromatin; cryo-EM; epigenetics; histone; kinetochore; microtubule spindle; mitosis; nucleosome.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interest
The authors declare no conflict of interest.
Figures




References
-
- Depinet TW, Zackowski JL, Earnshaw WC, Kaffe S, Sekhon GS, Stallard R, Sullivan BA, Vance GH, Van Dyke DL, Willard HF, et al. (1997). Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum. Mol Genet 6, 1195–1204. - PubMed
-
- Eichler EE (1999). Repetitive conundrums of centromere structure and function. Hum. Mol. Genet 8, 151–155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

