4-1BB Delineates Distinct Activation Status of Exhausted Tumor-Infiltrating CD8+ T Cells in Hepatocellular Carcinoma
- PMID: 31353502
- PMCID: PMC7154753
- DOI: 10.1002/hep.30881
4-1BB Delineates Distinct Activation Status of Exhausted Tumor-Infiltrating CD8+ T Cells in Hepatocellular Carcinoma
Abstract
Background and aims: Targeting costimulatory receptors with agonistic antibodies is a promising cancer immunotherapy option. We aimed to investigate costimulatory receptor expression, particularly 4-1BB (CD137 or tumor necrosis factor receptor superfamily member 9), on tumor-infiltrating CD8+ T cells (CD8+ tumor-infiltrating lymphocytes [TILs]) and its association with distinct T-cell activation features among exhausted CD8+ TILs in hepatocellular carcinoma (HCC).
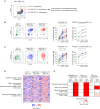
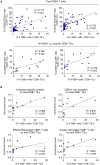
Approach and results: Tumor tissues, adjacent nontumor tissues, and peripheral blood were collected from HCC patients undergoing surgical resection (n = 79). Lymphocytes were isolated and used for multicolor flow cytometry, RNA-sequencing, and in vitro functional restoration assays. Among the examined costimulatory receptors, 4-1BB was most prominently expressed on CD8+ TILs. 4-1BB expression was almost exclusively detected on CD8+ T cells in the tumor-especially on programmed death 1 (PD-1)high cells and not PD-1int and PD-1neg cells. Compared to PD-1int and 4-1BBneg PD-1high CD8+ TILs, 4-1BBpos PD-1high CD8+ TILs exhibited higher levels of tumor reactivity and T-cell activation markers and significant enrichment for T-cell activation gene signatures. Per-patient analysis revealed positive correlations between percentages of 4-1BBpos cells among CD8+ TILs and levels of parameters of tumor reactivity and T-cell activation. Among highly exhausted PD-1high CD8+ TILs, 4-1BBpos cells harbored higher proportions of cells with proliferative and reinvigoration potential. Our 4-1BB-related gene signature predicted survival outcomes of HCC patients in the The Cancer Genome Atlas cohort. 4-1BB agonistic antibodies enhanced the function of CD8+ TILs and further enhanced the anti-PD-1-mediated reinvigoration of CD8+ TILs, especially in cases showing high levels of T-cell activation.
Conclusion: 4-1BB expression on CD8+ TILs represents a distinct activation state among highly exhausted CD8+ T cells in HCC. 4-1BB costimulation with agonistic antibodies may be a promising strategy for treating HCCs exhibiting prominent T-cell activation.
© 2019 The Authors. Hepatology published by Wiley Periodicals, Inc., on behalf of American Association for the Study of Liver Diseases.
Figures




References
-
- Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov 2018;8:1069‐1086. - PubMed
-
- Zhu AX, Finn RS, Edeline J, Cattan S, Ogasawara S, Palmer D, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE‐224): a non‐randomised, open‐label phase 2 trial. Lancet Oncol 2018;19:940‐952. - PubMed
-
- Mayes PA, Hance KW, Hoos A. The promise and challenges of immune agonist antibody development in cancer. Nat Rev Drug Discov 2018;17:509‐527. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

