STAT1 Gain-of-Function Mutations Cause High Total STAT1 Levels With Normal Dephosphorylation
- PMID: 31354696
- PMCID: PMC6635460
- DOI: 10.3389/fimmu.2019.01433
STAT1 Gain-of-Function Mutations Cause High Total STAT1 Levels With Normal Dephosphorylation
Abstract
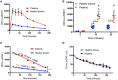
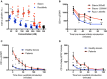
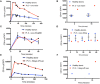
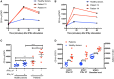
Signal transducer and activator of transcription (STAT1)1 gain of function (GOF) pathogenic variants have been associated with increased levels of phosphorylated STAT1 and STAT1-dependent cellular responses. Delayed dephosphorylation was proposed as the underlying mechanism leading to the characteristically raised pSTAT1 levels. We examined the levels of STAT1 protein and message as well as rates of STAT1 phosphorylation, dephosphorylation, and degradation associated with STAT1 GOF pathogenic variants. Fresh peripheral blood mononuclear cells (PBMC) from 14 STAT1 GOF patients carrying 10 different pathogenic variants in the coiled-coil, DNA binding, and SH2 domains and healthy donors were used to study STAT1 levels and phosphorylation (pSTAT1) following IFNγ and IFNα stimulation. STAT1 protein levels were measured by flow cytometry and immunoblot. STAT1 mRNA levels were measured using quantitative reverse transcription PCR. STAT1 protein degradation was studied using cycloheximide. Patient IFNγ and IFNα induced peak pSTAT1 was higher than in healthy controls. The velocity of pSTAT1 dephosphorylation after treatment of IFNγ stimulated CD14+ monocytes with the Janus Kinase (JAK)-inhibitor ruxolitinib was significantly faster in patient cells. STAT1 protein levels in patient CD14+ monocytes and CD3+ T cells were higher than in healthy donors. There was a strong and positive correlation between CD14+ STAT1 protein levels and peak pSTAT1 levels. Patient fresh PBMC STAT1 mRNA levels were increased at rest and after 16 h of incubation. STAT1 protein degradation was similar in patient and healthy volunteer cells. Patient IFNγ receptors 1 and 2 and JAK2 levels were normal. One patient in our cohort was treated with the oral JAK inhibitor ruxolitinib. Treatment was associated with normalization of both STAT1 protein and peak pSTAT1 levels. After JAK inhibitor treatment was stopped the patient's CD14+ monocyte STAT1 protein and peak phosphorylation levels increased proportionally. These findings suggest that patients with STAT1 GOF mutations have higher levels of total STAT1 protein, leading to high levels of pSTAT1 after stimulation, despite rapid STAT1 dephosphorylation and normal degradation.
Keywords: JAK inhibitors; STAT1; T cells; dephosphorylation; gain of function; mRNA; monocytes; protein.
Figures





References
-
- Uzel G, Sampaio EP, Lawrence MG, Hsu AP, Hackett M, Dorsey MJ, et al. . Dominant gain-of-function STAT1 mutations in FOXP3 wild-type immune dysregulation-polyendocrinopathy-enteropathy-X-linked-like syndrome. J Allergy Clin Immunol. (2013) 131:1611–23. 10.1016/j.jaci.2012.11.054 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

