Epigenetic Regulation Of Axon Regeneration and Glial Activation in Injury Responses
- PMID: 31354788
- PMCID: PMC6629966
- DOI: 10.3389/fgene.2019.00640
Epigenetic Regulation Of Axon Regeneration and Glial Activation in Injury Responses
Abstract
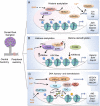
Injury to the nervous system triggers a multicellular response in which epigenetic mechanisms play an important role in regulating cell type-specific transcriptional changes. Here, we summarize recent progress in characterizing neuronal intrinsic and extrinsic chromatin reconfigurations and epigenetic changes triggered by axonal injury that shape neuroplasticity and glial functions. We specifically discuss regeneration-associated transcriptional modules comprised of transcription factors and epigenetic regulators that control axon growth competence. We also review epigenetic regulation of neuroinflammation and astroglial responses that impact neural repair. These advances provide a framework for developing epigenetic strategies to maximize adaptive alterations while minimizing maladaptive stress responses in order to enhance axon regeneration and achieve functional recovery after injury.
Keywords: CNS injury; axon regeneration; chromatin accessibility; epigenetic regulation; neural repair; neuroepigenetics; neuroinflammation; spinal cord injury.
Figures


References
-
- Beck K. D., Nguyen H. X., Galvan M. D., Salazar D. L., Woodruff T. M., Anderson A. J. (2010). Quantitative analysis of cellular inflammation after traumatic spinal cord injury: evidence for a multiphasic inflammatory response in the acute to chronic environment. Brain 133, 433–447. 10.1093/brain/awp322 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

