Extreme Diversity of Mycoviruses Present in Isolates of Rhizoctonia solani AG2-2 LP From Zoysia japonica From Brazil
- PMID: 31355150
- PMCID: PMC6640214
- DOI: 10.3389/fcimb.2019.00244
Extreme Diversity of Mycoviruses Present in Isolates of Rhizoctonia solani AG2-2 LP From Zoysia japonica From Brazil
Abstract
Zoysia japonica, in Brazil, is commonly infected by Rhizoctonia solani (R. solani) in humid and cool weather conditions. Eight isolates of R. solani, previously identified as belonging to the AG2-2 LP anastomosis group, isolated from samples from large path symptoms, were collected from three counties in São Paulo state (Brazil) and investigated for the presence of mycoviruses. After detection of double-strand RNA (dsRNA) in all samples, RNA_Seq analysis of ribosomal RNA-depleted total RNA from in vitro cultivated mycelia was performed. Forty-seven partial or complete viral unique RNA dependent-RNA polymerase (RdRp) sequences were obtained with a high prevalence of positive sense ssRNA viruses. Sequences were sufficiently different from the first match in BLAST searches suggesting that they all qualify as possible new viral species, except for one sequence showing an almost complete match with Rhizoctonia solani dsRNA virus 2, an alphapartitivirus. Surprisingly four large contigs of putative viral RNA could not be assigned to any existing clade of viruses present in the databases, but no DNA was detected corresponding to these fragments confirming their viral replicative nature. This is the first report on the occurrence of mycoviruses in R. solani AG2-2 LP in South America.
Keywords: Rhizoctonia solani; grass; multiple infection; mycoviruses; phylogenetic analysis; viral diversity; virus taxonomy.
Figures

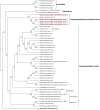


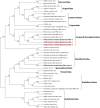
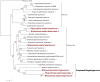


References
-
- Antoniolli D. (2015). “Produção, regularização e conquistas do mercado de gramas cultivadas no Brasil,” in Tópicos atuais em gramados IV, VII SIGRA – Simpósio sobre Gramados. FCA/UNESP. eds Mateus C.M.D'A., Villas Bôas R.L., Andrade T. F., Oliveira M.R., Backes C., Santos A.J.M., Godoy L.J.G. (Botucatu: Editora Fepaf, 9–22.
-
- Bharathan N., Saso H., Gudipati L., Bharathan S., Whited K. (2005). Double-stranded RNA: distribution and analysis among isolates of Rhizoctonia solani AG-2 to−13. Plant Pathol. 54, 196–203. 10.1111/j.1365-3059.2005.01159.x - DOI
-
- Chiba S., Salaipeth L., Lin Y. H. (2009). A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: molecular and biological characterization, taxonomic considerations, and potential for biological control. J. Virol. 3, 12801–12812. 10.1128/JVI.01830-09 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

