T cell receptor-based cancer immunotherapy: Emerging efficacy and pathways of resistance
- PMID: 31355495
- PMCID: PMC7027847
- DOI: 10.1111/imr.12772
T cell receptor-based cancer immunotherapy: Emerging efficacy and pathways of resistance
Abstract
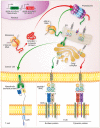
Adoptive cell transfer (ACT) using chimeric antigen receptor (CAR)-modified T cells can induce durable remissions in patients with refractory B-lymphoid cancers. By contrast, results applying CAR-modified T cells to solid malignancies have been comparatively modest. Alternative strategies to redirect T cell specificity and cytolytic function are therefore necessary if ACT is to serve a greater role in human cancer treatments. T cell receptors (TCRs) are antigen recognition structures physiologically expressed by all T cells that have complementary, and in some cases superior, properties to CARs. Unlike CARs, TCRs confer recognition to epitopes derived from proteins residing within any subcellular compartment, including the membrane, cytoplasm and nucleus. This enables TCRs to detect a broad universe of targets, such as neoantigens, cancer germline antigens, and viral oncoproteins. Moreover, because TCRs have evolved to efficiently detect and amplify antigenic signals, these receptors respond to epitope densities many fold smaller than required for CAR-signaling. Herein, we summarize recent clinical data demonstrating that TCR-based immunotherapies can mediate regression of solid malignancies, including immune-checkpoint inhibitor refractory cancers. These trials simultaneously highlight emerging mechanisms of TCR resistance. We conclude by discussing how TCR-based immunotherapies can achieve broader dissemination through innovations in cell manufacturing and non-viral genome integration techniques.
Keywords: CRISPR/Cas9; ImmTAC; TCR mimic; adoptive immunotherapy; genetic engineering.
© 2019 The Authors. Immunological Reviews Published by John Wiley & Sons Ltd.
Conflict of interest statement
Advisory/consulting: Aleta BioTherapeutics, Bellicum Pharmaceuticals, BMS, Cell Design Labs, G1 Therapeutics, Klus Pharma, Obsidian Therapeutics, Rxi Therapeutics (CAK); Honoraria: Kite/Gilead (CAK); Clinical research support: Kite/Gilead (CAK). CAK and SSC hold provisional patents related to T cell therapy and TCR engineering.
Figures




References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7‐34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

