Equine arteritis virus long-term persistence is orchestrated by CD8+ T lymphocyte transcription factors, inhibitory receptors, and the CXCL16/CXCR6 axis
- PMID: 31356622
- PMCID: PMC6692045
- DOI: 10.1371/journal.ppat.1007950
Equine arteritis virus long-term persistence is orchestrated by CD8+ T lymphocyte transcription factors, inhibitory receptors, and the CXCL16/CXCR6 axis
Abstract
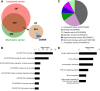
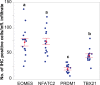
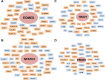
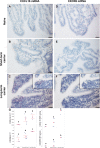
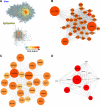
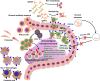
Equine arteritis virus (EAV) has the unique ability to establish long-term persistent infection in the reproductive tract of stallions and be sexually transmitted. Previous studies showed that long-term persistent infection is associated with a specific allele of the CXCL16 gene (CXCL16S) and that persistence is maintained despite the presence of local inflammatory and humoral and mucosal antibody responses. Here, we performed transcriptomic analysis of the ampullae, the primary site of EAV persistence in long-term EAV carrier stallions, to understand the molecular signatures of viral persistence. We demonstrated that the local CD8+ T lymphocyte response is predominantly orchestrated by the transcription factors eomesodermin (EOMES) and nuclear factor of activated T-cells cytoplasmic 2 (NFATC2), which is likely modulated by the upregulation of inhibitory receptors. Most importantly, EAV persistence is associated with an enhanced expression of CXCL16 and CXCR6 by infiltrating lymphocytes, providing evidence of the implication of this chemokine axis in the pathogenesis of persistent EAV infection in the stallion reproductive tract. Furthermore, we have established a link between the CXCL16 genotype and the gene expression profile in the ampullae of the stallion reproductive tract. Specifically, CXCL16 acts as a "hub" gene likely driving a specific transcriptional network. The findings herein are novel and strongly suggest that RNA viruses such as EAV could exploit the CXCL16/CXCR6 axis in order to modulate local inflammatory and immune responses in the male reproductive tract by inducing a dysfunctional CD8+ T lymphocyte response and unique lymphocyte homing in the reproductive tract.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142(3):629–33. - PubMed
-
- Balasuriya U, MacLachlan NJ. Equine Viral Arteritis In: Sellon DC, Long MT, editors. Equine Infectious Diseases. 2nd ed St. Louis, MO: Saunders; 2013. p. 169–81. 10.1016/S1473-3099(13)70227-5 - DOI
-
- Balasuriya UBR, Carossino M, Timoney PJ. Equine viral arteritis: a respiratory and reproductive disease of significant economic importance to the equine industry. Equine Vet Educ. 2016;In press. 10.1111/eve.12566 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

