Diminished Condensin Gene Expression Drives Chromosome Instability That May Contribute to Colorectal Cancer Pathogenesis
- PMID: 31357676
- PMCID: PMC6721357
- DOI: 10.3390/cancers11081066
Diminished Condensin Gene Expression Drives Chromosome Instability That May Contribute to Colorectal Cancer Pathogenesis
Abstract
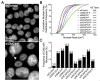
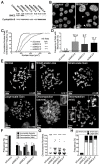
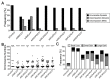
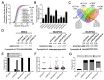
Chromosome instability (CIN), or constantly evolving chromosome complements, is a form of genome instability implicated in the development and progression of many cancer types, however, the molecular determinants of CIN remain poorly understood. Condensin is a protein complex involved in chromosome compaction, and recent studies in model organisms show that aberrant compaction adversely impacts mitotic fidelity. To systematically assess the clinical and fundamental impacts that reduced condensin gene expression have in cancer, we first assessed gene copy number alterations of all eight condensin genes. Using patient derived datasets, we show that shallow/deep deletions occur frequently in 12 common cancer types. Furthermore, we show that reduced expression of each gene is associated with worse overall survival in colorectal cancer patients. To determine the overall impact that reduced condensin gene expression has on CIN, a comprehensive siRNA-based screen was performed in two karyotypically stable cell lines. Following gene silencing, quantitative imaging microscopy identified increases in CIN-associated phenotypes, including changes in nuclear areas, micronucleus formation, and chromosome numbers. Although silencing corresponded with increases in CIN phenotypes, the most pronounced phenotypes were observed following SMC2 and SMC4 silencing. Collectively, our clinical and fundamental findings suggest reduced condensin expression and function may be a significant, yet, underappreciated driver of colorectal cancer.
Keywords: chromosome instability; colorectal cancer; condensin; micronucleus; single cell quantitative imaging microscopy.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- American Cancer Society . Cancer Facts Figures 2019. American Cancer Society; Atlanta, GA, USA: 2019.
-
- Canadian Cancer Society’s Advisory Committee on Cancer Statistics . Canadian Cancer Statistics 2017. Canadian Cancer Society; Toronto, ON, Canada: 2017.
Grants and funding
LinkOut - more resources
Full Text Sources

