Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma
- PMID: 31358576
- PMCID: PMC6872444
- DOI: 10.1136/gutjnl-2019-318882
Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma
Abstract
Objective: The lack of highly sensitive and specific diagnostic biomarkers is a major contributor to the poor outcomes of patients with hepatocellular carcinoma (HCC). We sought to develop a non-invasive diagnostic approach using circulating cell-free DNA (cfDNA) for the early detection of HCC.
Design: Applying the 5hmC-Seal technique, we obtained genome-wide 5-hydroxymethylcytosines (5hmC) in cfDNA samples from 2554 Chinese subjects: 1204 patients with HCC, 392 patients with chronic hepatitis B virus infection (CHB) or liver cirrhosis (LC) and 958 healthy individuals and patients with benign liver lesions. A diagnostic model for early HCC was developed through case-control analyses using the elastic net regularisation for feature selection.
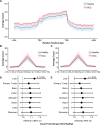
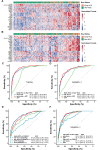
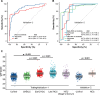
Results: The 5hmC-Seal data from patients with HCC showed a genome-wide distribution enriched with liver-derived enhancer marks. We developed a 32-gene diagnostic model that accurately distinguished early HCC (stage 0/A) based on the Barcelona Clinic Liver Cancer staging system from non-HCC (validation set: area under curve (AUC)=88.4%; (95% CI 85.8% to 91.1%)), showing superior performance over α-fetoprotein (AFP). Besides detecting patients with early stage or small tumours (eg, ≤2.0 cm) from non-HCC, the 5hmC model showed high capacity for distinguishing early HCC from high risk subjects with CHB or LC history (validation set: AUC=84.6%; (95% CI 80.6% to 88.7%)), also significantly outperforming AFP. Furthermore, the 5hmC diagnostic model appeared to be independent from potential confounders (eg, smoking/alcohol intake history).
Conclusion: We have developed and validated a non-invasive approach with clinical application potential for the early detection of HCC that are still surgically resectable in high risk individuals.
Keywords: cancer; hepatobiliary cancer; hepatocellular carcinoma.
© Author(s) (or their employer(s)) 2019. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: The 5hmC-Seal technology was invented by CH and was licensed by Shanghai Epican Genetech Co. Ltd. for clinical applications in human diseases from the University of Chicago. XL is a co-founder of Shanghai Epican Genetech Co. Ltd. CH and WZ are shareholders of Shanghai Epican Genetech Co. Ltd. CH is a scientific founder of Accent Therapeutics, Inc. and a member of its scientific advisory board. All other authors report no potential conflicts of interest.
Figures



Comment in
-
New warning signs on the road: 5-hydroxymethylcytosine-based liquid biopsy for the early detection of hepatocellular carcinoma.Gut. 2019 Dec;68(12):2103-2104. doi: 10.1136/gutjnl-2019-319339. Epub 2019 Aug 7. Gut. 2019. PMID: 31391197 No abstract available.
-
Are the 5-hydroxymethylcytosine-based wd-scores really superior over α-fetoprotein for the early diagnosis of hepatocellular carcinoma?Gut. 2020 Oct;69(10):1892. doi: 10.1136/gutjnl-2019-319853. Epub 2019 Oct 25. Gut. 2020. PMID: 31653786 No abstract available.
-
Reply to 'Are the 5-hydroxymethylcytosine-based wd-scores really superior over α-fetoprotein for the early diagnosis of hepatocellular carcinoma?'.Gut. 2020 Oct;69(10):1903-1904. doi: 10.1136/gutjnl-2019-320298. Epub 2019 Dec 23. Gut. 2020. PMID: 31871104 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
