Genome of the Komodo dragon reveals adaptations in the cardiovascular and chemosensory systems of monitor lizards
- PMID: 31358948
- PMCID: PMC6668926
- DOI: 10.1038/s41559-019-0945-8
Genome of the Komodo dragon reveals adaptations in the cardiovascular and chemosensory systems of monitor lizards
Abstract
Monitor lizards are unique among ectothermic reptiles in that they have high aerobic capacity and distinctive cardiovascular physiology resembling that of endothermic mammals. Here, we sequence the genome of the Komodo dragon Varanus komodoensis, the largest extant monitor lizard, and generate a high-resolution de novo chromosome-assigned genome assembly for V. komodoensis using a hybrid approach of long-range sequencing and single-molecule optical mapping. Comparing the genome of V. komodoensis with those of related species, we find evidence of positive selection in pathways related to energy metabolism, cardiovascular homoeostasis, and haemostasis. We also show species-specific expansions of a chemoreceptor gene family related to pheromone and kairomone sensing in V. komodoensis and other lizard lineages. Together, these evolutionary signatures of adaptation reveal the genetic underpinnings of the unique Komodo dragon sensory and cardiovascular systems, and suggest that selective pressure altered haemostasis genes to help Komodo dragons evade the anticoagulant effects of their own saliva. The Komodo dragon genome is an important resource for understanding the biology of monitor lizards and reptiles worldwide.
Conflict of interest statement
Competing Interests: The authors have no competing interests to declare
Figures


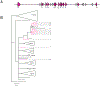
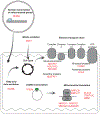
Comment in
-
Decoding dragon DNA.Nat Rev Genet. 2019 Oct;20(10):564-565. doi: 10.1038/s41576-019-0168-5. Nat Rev Genet. 2019. PMID: 31388141 No abstract available.
References
-
- Chapman AD Numbers of Living Species in Australia and the World (Australian Biological Resources Study, 2009).
-
- Collar DC, Schulte JA & Losos JB Evolution of extreme body size disparity in monitor lizards (Varanus). Evolution (N. Y). 65, 2664–2680 (2011). - PubMed
-
- Jensen B, Wang T, Christoffels VM & Moorman AFM Evolution and development of the building plan of the vertebrate heart. Biochim. Biophys. Acta - Mol. Cell Res 1833, 783–794 (2013). - PubMed
-
- Auffenberg W The Behavioral Ecology of the Komodo Monitor (University Presses of Florida, 1981).
-
- Green B, King D, Braysher M & Saim A Thermoregulation, water turnover and energetics of free-living komodo dragons, Varanus komodoensis. Comp. Biochem. Physiol. Part A Physiol 99, 97–101 (1991).

