Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans
- PMID: 31358989
- PMCID: PMC6953633
- DOI: 10.1038/s41593-019-0447-7
Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans
Abstract
Post-traumatic stress disorder (PTSD) is a major problem among military veterans and civilians alike, yet its pathophysiology remains poorly understood. We performed a genome-wide association study and bioinformatic analyses, which included 146,660 European Americans and 19,983 African Americans in the US Million Veteran Program, to identify genetic risk factors relevant to intrusive reexperiencing of trauma, which is the most characteristic symptom cluster of PTSD. In European Americans, eight distinct significant regions were identified. Three regions had values of P < 5 × 10-10: CAMKV; chromosome 17 closest to KANSL1, but within a large high linkage disequilibrium region that also includes CRHR1; and TCF4. Associations were enriched with respect to the transcriptomic profiles of striatal medium spiny neurons. No significant associations were observed in the African American cohort of the sample. Results in European Americans were replicated in the UK Biobank data. These results provide new insights into the biology of PTSD in a well-powered genome-wide association study.
Figures


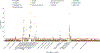
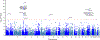

References
-
- Fulton JJ et al. The prevalence of posttraumatic stress disorder in Operation Enduring Freedom/Operation Iraqi Freedom (OEF/OIF) Veterans: a meta-analysis. J Anxiety Disord 31, 98–107 (2015). - PubMed
Methods References
-
- Gaziano JM et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214–23 (2016). - PubMed

