How to get away with nonsense: Mechanisms and consequences of escape from nonsense-mediated RNA decay
- PMID: 31359616
- PMCID: PMC10685860
- DOI: 10.1002/wrna.1560
How to get away with nonsense: Mechanisms and consequences of escape from nonsense-mediated RNA decay
Abstract
Nonsense-mediated RNA decay (NMD) is an evolutionarily conserved RNA quality control process that serves both as a mechanism to eliminate aberrant transcripts carrying premature stop codons, and to regulate expression of some normal transcripts. For a quality control process, NMD exhibits surprising variability in its efficiency across transcripts, cells, tissues, and individuals in both physiological and pathological contexts. Whether an aberrant RNA is spared or degraded, and by what mechanism, could determine the phenotypic outcome of a disease-causing mutation. Hence, understanding the variability in NMD is not only important for clinical interpretation of genetic variants but also may provide clues to identify novel therapeutic approaches to counter genetic disorders caused by nonsense mutations. Here, we discuss the current knowledge of NMD variability and the mechanisms that allow certain transcripts to escape NMD despite the presence of NMD-inducing features. This article is categorized under: RNA Turnover and Surveillance > Turnover/Surveillance Mechanisms RNA in Disease and Development > RNA in Disease RNA Turnover and Surveillance > Regulation of RNA Stability.
Keywords: RNA processing; mRNA surveillance; nonsense-mediated RNA decay; translation.
© 2019 Wiley Periodicals, Inc.
Figures

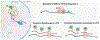
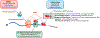

References
-
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, and Jacobson A. 2004. A faux 3’-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 432:112–118. - PubMed
-
- Amrani N, and Jacobson A. 2013. All termination events are not equal: premature termination in yeast is aberrant and triggers NMD. In Madame Curie Bioscience Database. Landes Bioscience, Austin, TX.
-
- Anczukow O, Ware MD, Buisson M, Zetoune AB, Stoppa-Lyonnet D, Sinilnikova OM, and Mazoyer S. 2008. Does the nonsense-mediated mRNA decay mechanism prevent the synthesis of truncated BRCA1, CHK2, and p53 proteins? Hum Mutat. 29:65–73. - PubMed
-
- Asiful Islam M, Alam F, Kamal MA, Gan SH, Wong KK, and Sasongko TH. 2017. Therapeutic Suppression of Nonsense Mutation: An Emerging Target in Multiple Diseases and Thrombotic Disorders. Curr Pharm Des. 23:1598–1609. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

